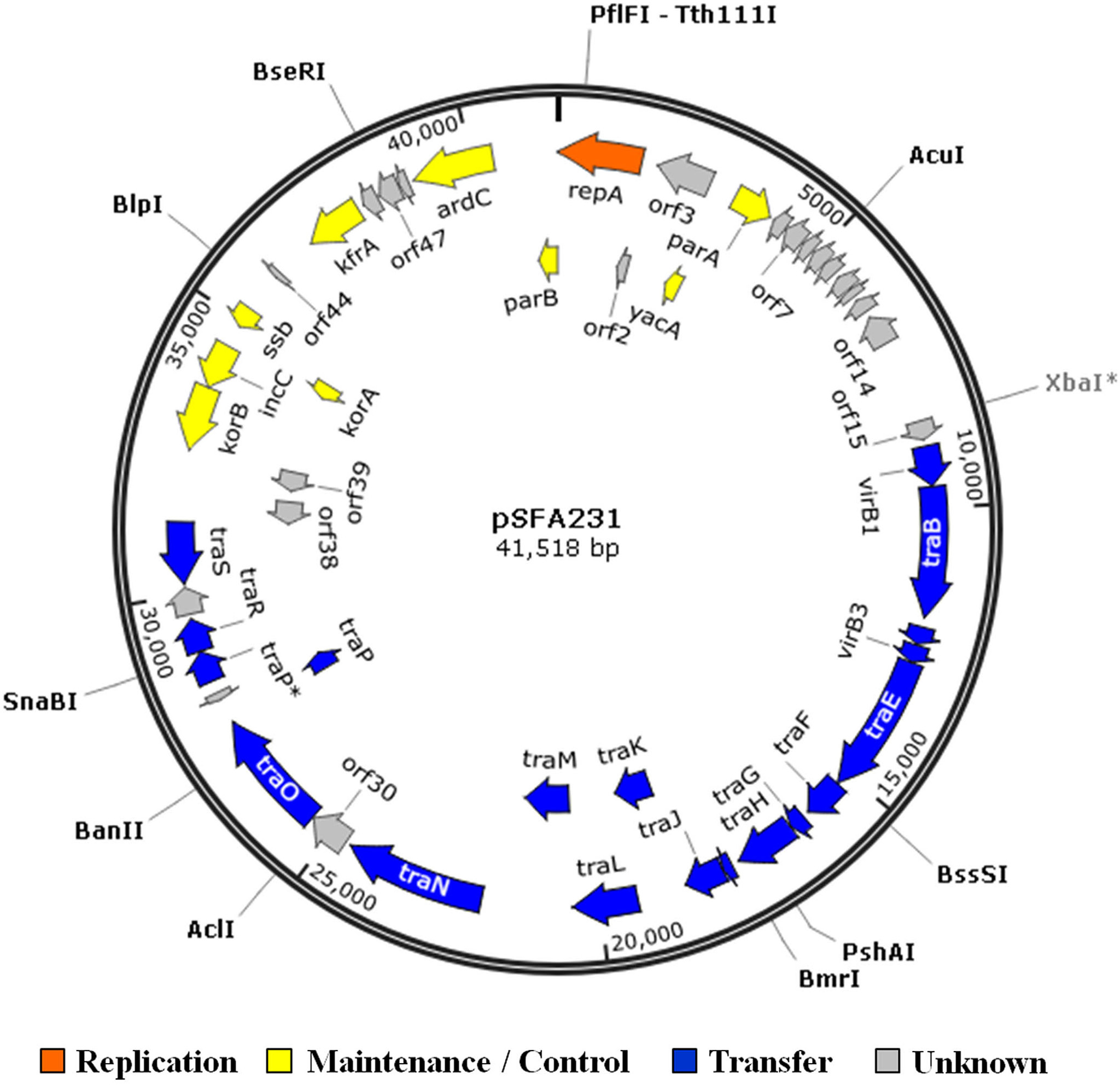
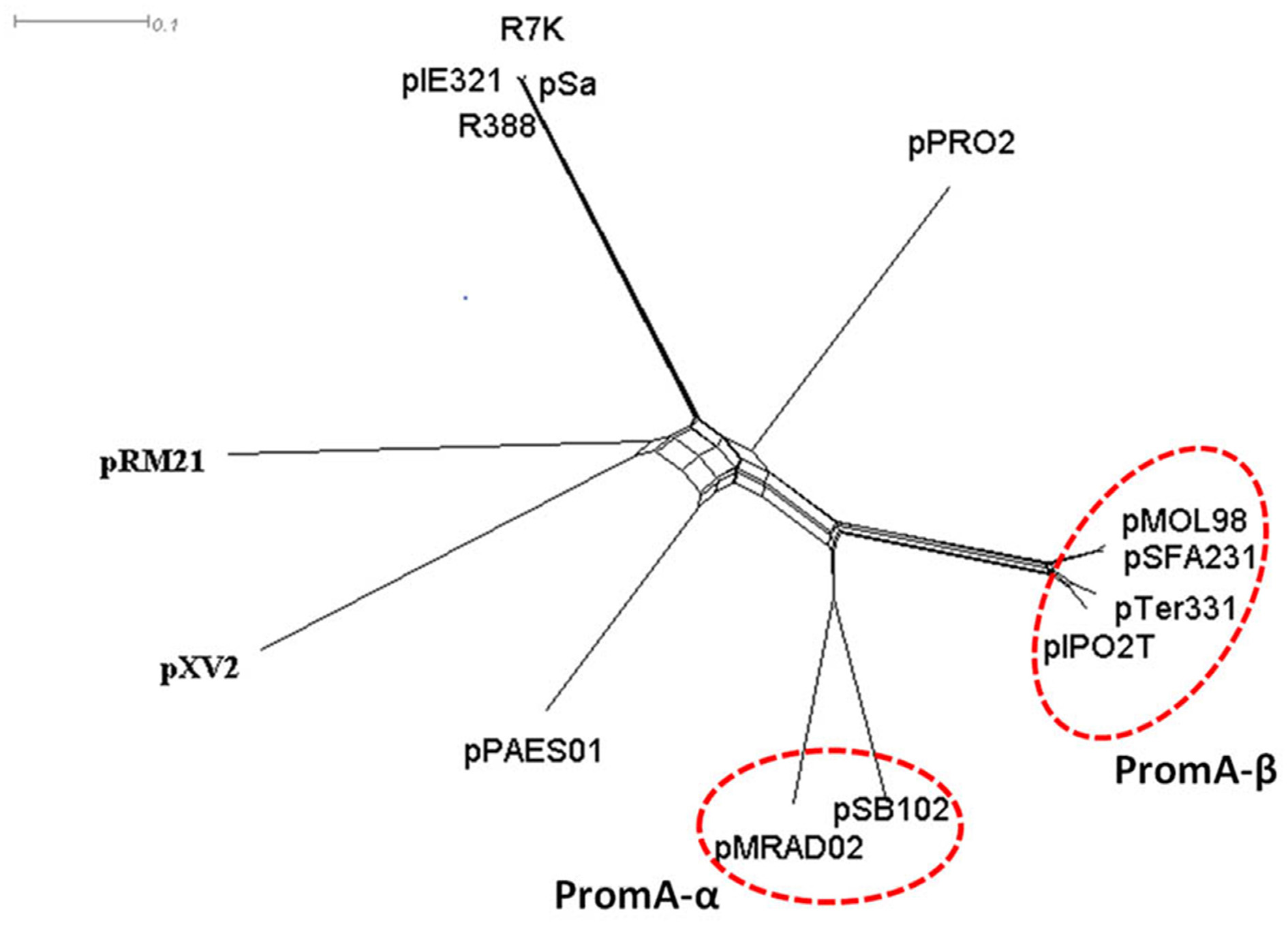
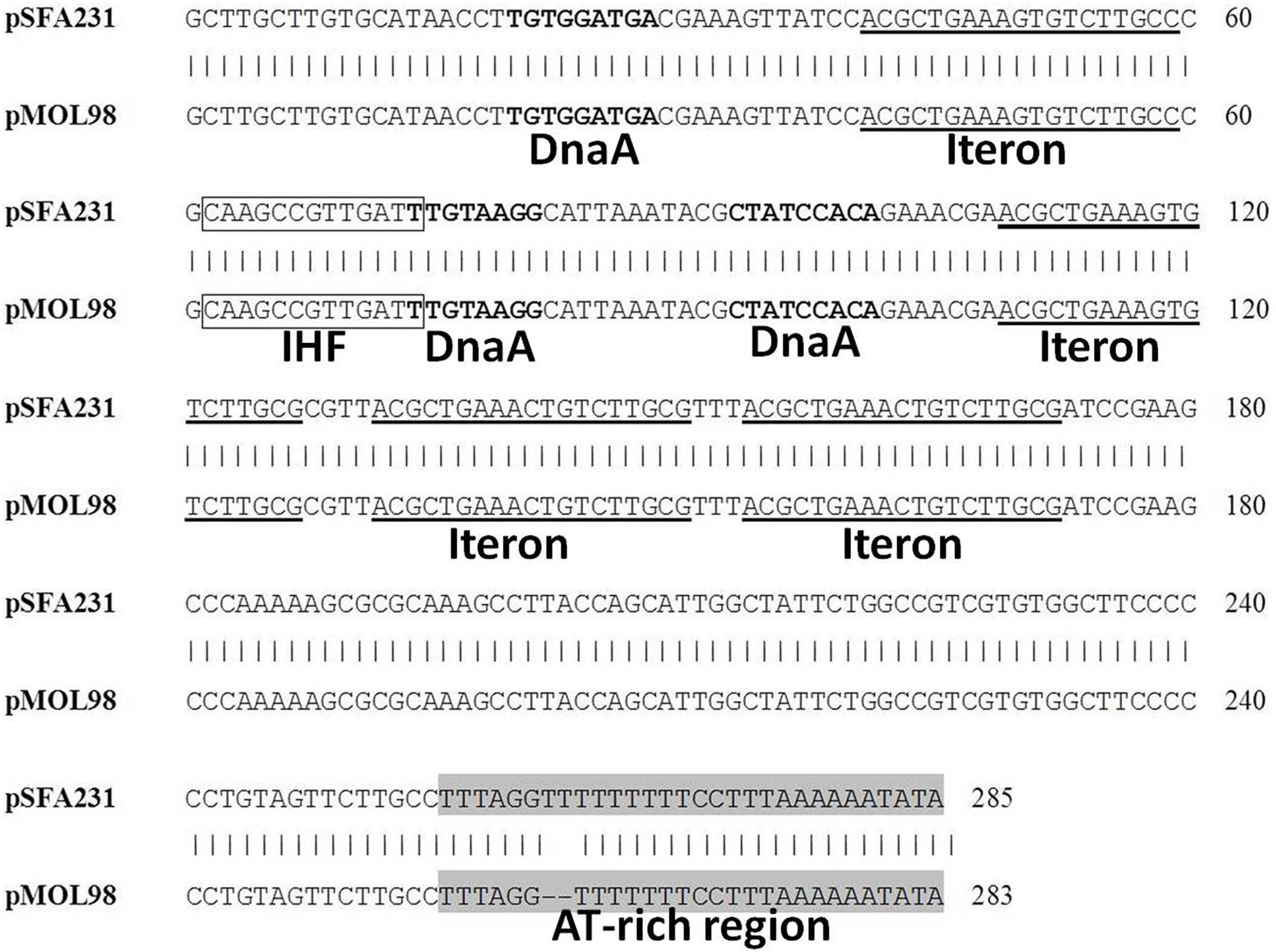
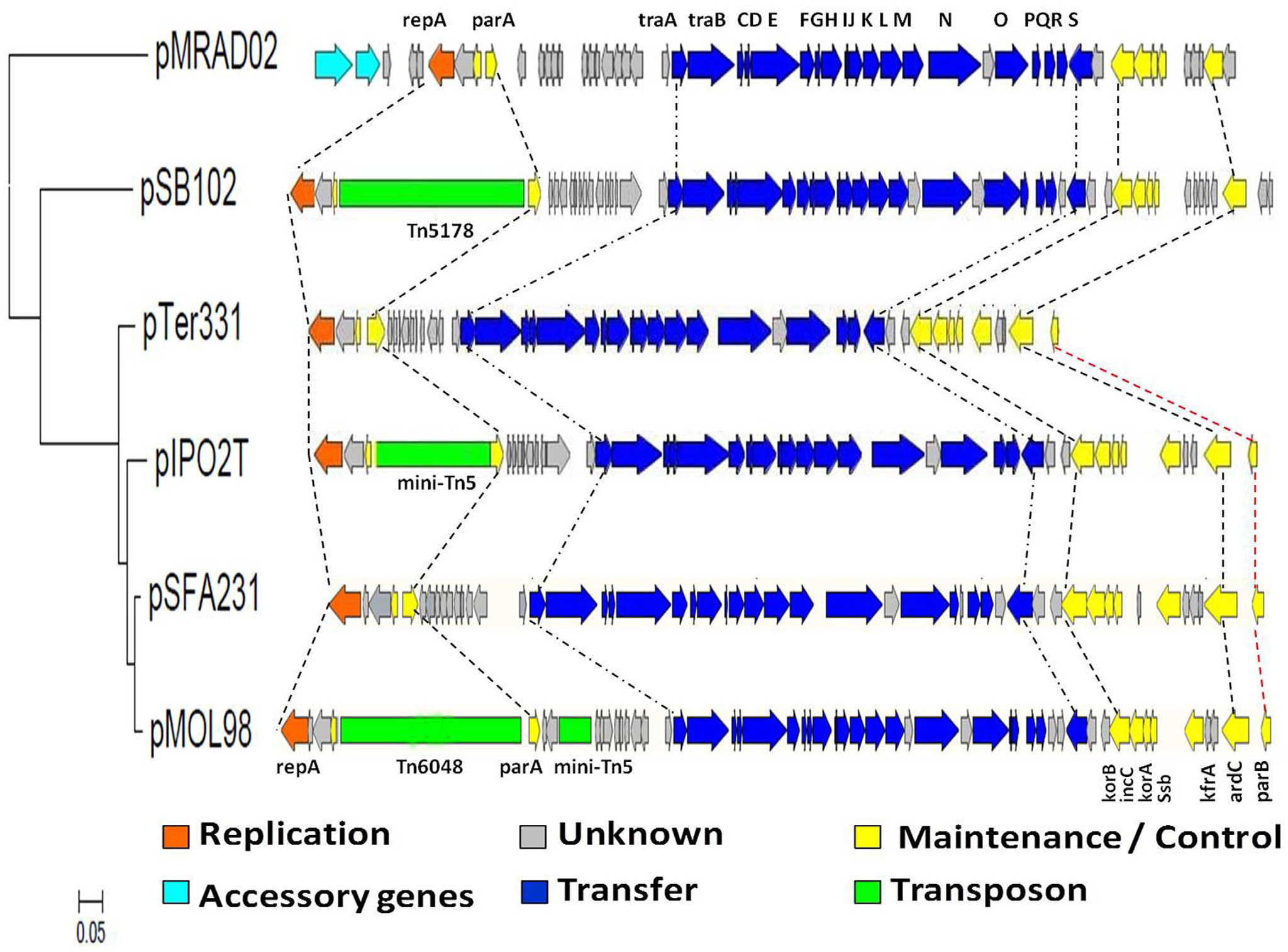
Begre s, koenig t
CURRICULUM VITAE (aktualisiert 20.05.2012) Name Berufliche Qualifikation Facharzt für Psychiatrie und Psychotherapie FMH FMH Schwerpunkt Konsiliar- und Liaisonpsychiatrie Facharzt für Innere Medizin FMH Fähigkeitsausweis für Psychosomatische und Psychosoziale Fähigkeitsausweis für Delegierte Psychotherapie FMPP Leiter FMH-Weiterbildungsstätte Psychiatrie und Psychotherapie,