Kamagra gibt es auch als Kautabletten, die sich schneller auflösen als normale Pillen. Manche Patienten empfinden das als angenehmer. Wer sich informieren will, findet Hinweise unter kamagra kautabletten.
Ews-fli1-mediated suppression of the ras-antagonist sprouty 1 (spry1) confers aggressiveness to ewing sarcoma
Oncogene (2016), 1–11 2016 Macmillan Publishers Limited All rights reserved 0950-9232/16
ORIGINAL ARTICLEEWS-FLI1-mediated suppression of the RAS-antagonistSprouty 1 (SPRY1) confers aggressiveness to Ewing sarcoma
F Cidre-Aranaz1, TGP Grünewald2,3, D Surdez2, L García-García1, J Carlos Lázaro1, T Kirchner3, L González-González1, A Sastre4,P García-Miguel4, SE López-Pérez1, S Monzón1,5, O Delattre2 and J Alonso1
Ewing sarcoma is characterized by chromosomal translocations fusing the EWS gene with various members of the ETS familyof transcription factors, most commonly FLI1. EWS-FLI1 is an aberrant transcription factor driving Ewing sarcoma tumorigenesisby either transcriptionally inducing or repressing specific target genes. Herein, we showed that Sprouty 1 (SPRY1), which is aphysiological negative feedback inhibitor downstream of fibroblast growth factor (FGF) receptors (FGFRs) and other RAS-activatingreceptors, is an EWS-FLI1 repressed gene. EWS-FLI1 knockdown specifically increased the expression of SPRY1, while other Sproutyfamily members remained unaffected. Analysis of SPRY1 expression in a panel of Ewing sarcoma cells showed that SPRY1 wasnot expressed in Ewing sarcoma cell lines, suggesting that it could act as a tumor suppressor gene in these cells. In agreement,induction of SPRY1 in three different Ewing sarcoma cell lines functionally impaired proliferation, clonogenic growth and migration.
In addition, SPRY1 expression inhibited extracellular signal-related kinase/mitogen-activated protein kinase (MAPK) signalinginduced by serum and basic FGF (bFGF). Moreover, treatment of Ewing sarcoma cells with the potent FGFR inhibitor PD-173074reduced bFGF-induced proliferation, colony formation and in vivo tumor growth in a dose-dependent manner, thus mimickingSPRY1 activity in Ewing sarcoma cells. Although the expression of SPRY1 was low when compared with other tumors, SPRY1was variably expressed in primary Ewing sarcoma tumors and higher expression levels were significantly associated with improvedoutcome in a large patient cohort. Taken together, our data indicate that EWS-FLI1-mediated repression of SPRY1 leads tounrestrained bFGF-induced cell proliferation, suggesting that targeting the FGFR/MAPK pathway can constitute a promisingtherapeutic approach for this devastating disease.
Oncogene advance online publication, 4 July 2016;
Here we report on the tumor suppressive role of another
Ewing sarcomas are aggressive bone and soft-tissue sarcomas
repressed EWS-FLI1-targeted gene, namely Sprouty 1 (SPRY1),
mostly affecting children and young adults.Although the 5-year
which is a negative feedback inhibitor of the RAS/mitogen-
survival in patients with localized disease increased significantly
activated protein kinase/extracellular signal-related kinase
on the addition of systemic chemotherapy to protocol treatments
(RAS/MAPK/ERK) pathway downstream of the fibroblast growth
in the 70–80 the prognosis and survival of patients with
factor receptor (FGFR).
metastatic or recurrent disease remained generally very poor.
SPRY1 is part of the mammalian Sprouty gene family consisting
Indeed, Ewing sarcoma features high rates of early metastasis with
of four members (SPRY1–4), which share important sequence
20% of patients having detectable metastasis at diagnosis.
similaritiessuch as a highly conserved cysteine-rich domain
The molecular hallmarks of Ewing sarcoma are nonrandom
in the C-terminal region (which is also found in the SPRED family
chromosomal translocations generating in-frame fusion of the
of proteins) and a short amino acid sequence in the N-terminus.
EWS gene on chromosome 22 and the C-terminus of a member
SPRY proteins differ largely in their tissue distribution, activity
of the ETS family of transcription factors (that is, FLI1, ERG, ETV1, FEV,
and interaction partners,thus suggesting non-redundant
ETV4 and POU5F1) including the DNA-binding domain(reviewed in
functions. SPRY1 is an upstream antagonist of RAS that is
Mackintosh et This fusion gives rise to aberrant EWS-ETS
activated by ERK, providing a negative feedback loop for RAS
transcription factors, EWS-FLI1 being present in 85% of cases.
signaling. Of note, about one-third of all human cancers are
EWS-FLI1 induces massive deregulation of protein expression
thought to carry a mutated RAS gene that activates downstream
by either transcriptionally inducing or repressing specific target
It has been suggested that SPRY1 may have a tumor
genes, many of which are involved in the oncogenic process.
suppressor function in specific tumors, as its expression is
For instance, EWS-FLI1 induces the expression of NR0B1 (DAX1),
decreased in several human cancers such as breast and prostate
EGR2, NKX2.2, CCK, PRKCB or STEAP1,while suppressing IGFBP3,
LOX, DKK1 or TGFBIIR.All these genes have been shown to
overexpression in tumor cell lines inhibits cell proliferation,
be important in Ewing sarcoma pathogenesis.
migration and anchorage-independent growth in vitro
1Unidad de Tumores Sólidos Infantiles, Instituto de Investigación de Enfermedades Raras, Instituto de Salud Carlos III, Majadahonda, Spain; 2INSERM U830 ‘Genetics and Biologyof Cancers' Institut Curie Research Center, Paris, France; 3Laboratory for Pediatric Sarcoma Biology, Institute of Pathology, LMU Munich, Munich, Germany; 4Unidad hemato-oncología pediátrica, Hospital Infantil Universitario La Paz, Madrid, Spain and 5Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER U758), Instituto deSalud Carlos III, Madrid, Spain. Correspondence: Dr J Alonso, Unidad de Tumores Sólidos Infantiles, Instituto de Investigación de Enfermedades Raras, Instituto de Salud Carlos III,Ctra. Majadahonda-Pozuelo km 2, Majadahonda Madrid 28220, Spain.
E-mail: Received 5 January 2016; revised 5 May 2016; accepted 30 May 2016

SPRY1 is a tumor suppressor in Ewing sarcoma
F Cidre-Aranaz et al
In this study, we show that SPRY1 acts as a tumor suppressor in
A673/TR/shEF grown in the absence of doxycycline. However,
Ewing sarcoma cells, and that SPRY1 repression is necessary for cell
a strong induction of SPRY1 protein was observed on doxycycline-
proliferation and migration. Interestingly, SPRY1 repression was
mediated EWS-FLI1 knockdown.
important to ERK pathway activation. Moreover, FGFR inhibition
We next studied whether the inhibition of SPRY1 expression
mimicked SPRY1 effect on proliferation and growth, indicating that
could be a common feature of Ewing sarcoma cells. We first
SPRY1 has an important role in Ewing sarcoma. Finally, elevated
analyzed the levels of SPRY1 mRNA and protein in a panel of eight
SPRY1 expression correlated with improved overall survival of
Ewing sarcoma cell lines harboring different EWS-FLI1 or EWS-ERG
Ewing sarcoma patients and inversely correlated with metastasis
fusion proteins (Supplementary Table 1). As shown in
at diagnosis. Collectively, our data indicate that EWS-FLI1-mediated
SPRY1 mRNA and protein were undetectable in all Ewing
repression of SPRY1 confers a growth advantage to Ewing sarcoma
sarcoma cell lines analyzed. Interestingly, the mRNA levels of
cells, and that SPRY1 levels constitute a novel biomarker for outcome
the other members of the SPRY family were variably expressed in
prediction of Ewing sarcoma patients. Taken together, these results
this panel of Ewing sarcoma cells. These data could also be
suggest a rationale for targeting FGFR/SPRY1/RAS/MAPK/ERK
confirmed assessing larger public data sets. For instance, analysis
pathway as a new therapeutic approach in this devastating disease.
of Cancer Cell Line Encyclopedia data setshowed that Ewing sarcoma cell linesexhibited the lowest SPRY1 levels among all tumor cell lines
analyzed (Supplementary Figure 1).
SPRY1 expression is strongly inhibited by EWS-FLI1 in Ewingsarcoma cell lines
SPRY1 induction impairs cell proliferation of Ewing sarcoma cells
Analysis of a gene expression profile of A673 Ewing sarcoma cell
The strong downregulation of SPRY1 by EWS-FLI1, its absence of
line genetically modified to express a specific small hairpin RNA
expression in Ewing sarcoma cell lines and the finding that it acts
directed against EWS-FLI1 mRNA on doxycycline stimulation (A673/
as a negative feedback inhibitor of the RAS/MAPK/ERK cascade
TR/shEF) (Gene Expression Omnibus accession code: GSE36007)
suggest a potential function of SPRY1 inhibition in Ewing sarcoma.
indicated that SPRY1 is strongly downregulated by EWS-FLI1. These
To test this hypothesis, we generated three doxycycline-inducible
microarray results were confirmed by reverse transcription–quanti-
SPRY1 Ewing sarcoma cell lines (A673/TR/SPRY1, SKES/TR/SPRY1
tative PCR experiments. As depicted in EWS-FLI1
and SKNMC/TR/SPRY1) and subjected them to several functional
knockdown led to a dramatic re-expression of SPRY1 mRNA (up to
assays. As shown in these genetically modified Ewing
1000-fold compared with controls), whereas the mRNA levels
cell lines express high levels of SPRY1 protein on doxycycline
of the other members of the SPRY family (SPRY2, 3 and 4) were
stimulation, whereas the levels of the EWS-FLI1 oncoprotein
only minimally affected. Analysis of SPRY1 protein levels in the
remain unaffected. Thus, they constitute a suitable model to test
A673/TR/shEF cell model confirmed these results. As shown in
the consequences of exclusive SPRY1 re-expression in Ewing
, SPRY1 protein was undetectable by western blotting in
sarcoma without affecting the levels of EWS-FLI1 oncoprotein.
Figure 1. SPRY1 is negatively regulated by EWS-FLI1 oncoprotein. (a) Time course of SPRY1, 2, 3 and 4 on EWS-FLI1 doxycycline-inducible
knockdown in A673/TR/shEF. EWS-FLI1 expression and two known target genes such as LOX and NR0B1 were included as controls. mRNAlevels were quantified by real-time reverse transcription–quantitative PCR (RT–qPCR), normalized to that of TBP (reference gene) and referred
to unstimulated cells. Figure shows data of one out of three independent experiments done in triplicate with equivalent results. EWS-FLI1inhibition in A673/TR/shEF cells selectively upregulates SPRY1 more than 1000 times over the rest of the members of the SPRY family of genes.
As expected, LOX appears upregulatedand NR0B1 downregulatedon EWS-FLI1 knockdown. (b) SPRY1 protein is re-expressed on EWS-FLI1knockdown in A673/TR/shEF cells. SPRY1 protein is undetectable by western blotting in A673 cells grown in the absence of doxycyclineand thus expressing EWS-FLI1. Incubation of A673/TR/shEWSFLI1 cells with doxycycline (1 μg/ml, 72 h) inhibits EWS-FLI1 expression and
dramatically induces re-expression of SPRY1 protein. Tubulin was used as a control for loading and transferring. (c) SPRY1 is undetectable
at protein level by western blotting in eight Ewing sarcoma cell lines. Expression of the different EWS-ETS proteins is also shown. Tubulin wasused as a control for loading and transferring. (d) SPRY1, 2, 3 and 4 mRNA levels in Ewing sarcoma cell lines. Box plot shows the absence
of SPRY1 expression in all Ewing sarcoma cell lines tested relative to other members of the SPRY family. The figure shows the expression levelsnormalized to that of TBP (reference gene).
Oncogene (2016) 1 – 11
2016 Macmillan Publishers Limited
SPRY1 is a tumor suppressor in Ewing sarcomaF Cidre-Aranaz et al
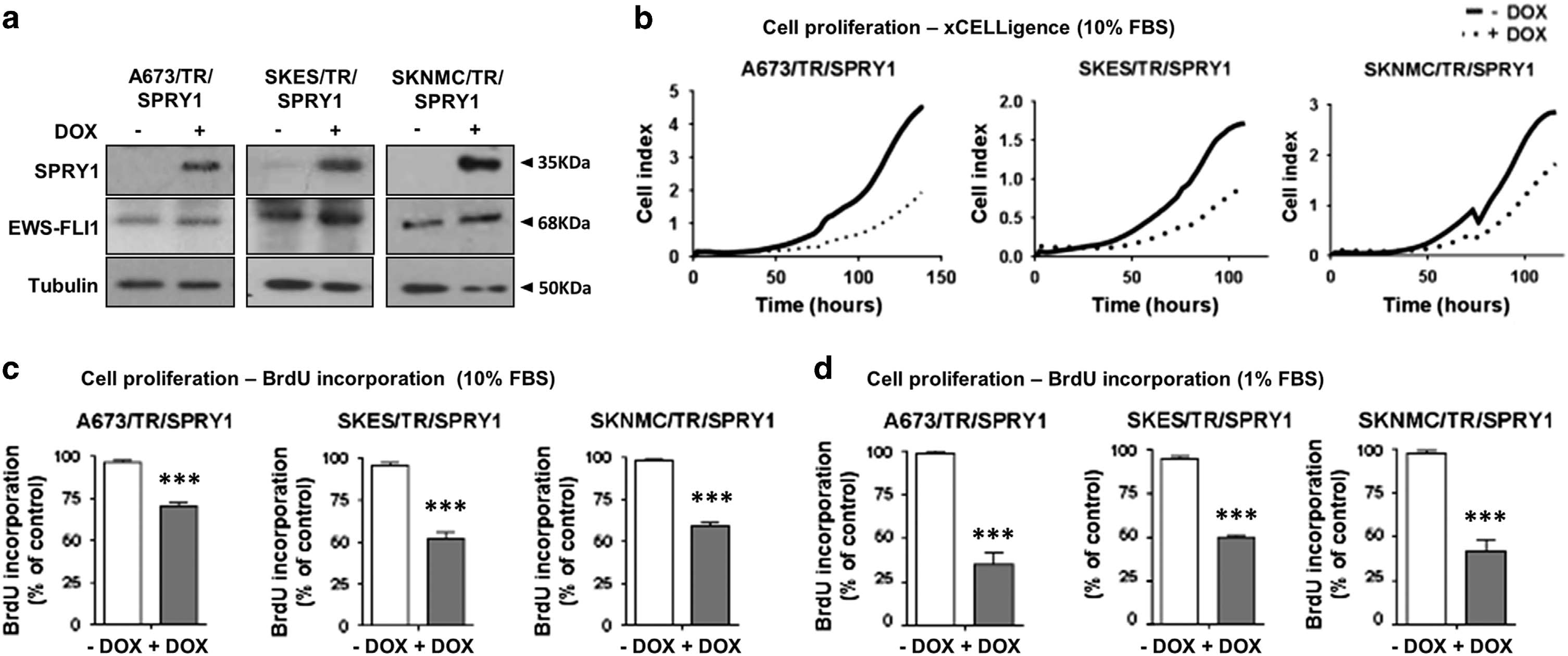
Figure 2. SPRY1 re-expression impairs proliferation in Ewing sarcoma cell lines. (a) A673/TR, SKES/TR and SKNMC/TR Ewing cell lines
expressing constitutively the tetracycline repressor (TR) were infected with a doxycycline-inducible lentiviral vector encoding theSPRY1 cDNA. The figure shows the expression of SPRY1 protein in whole protein extracts isolated from A673/TR/SPRY1 (clone 1), SKES/TR/
SPRY1 (clone 7) and SKNMC/TR/SPRY1 (clone 2) cells stimulated with doxycycline (DOX, 1 μg/ml, 72 h). High SPRY1 levels were detected in
all three cell lines after doxycycline stimulation. EWS-FLI1 expression was not affected by SPRY1 ectopic expression. The same blotwas stripped and incubated with anti-tubulin as a control for loading and transferring. (b) Cell proliferation was assayed in A673/TR/SPRY1,
SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells using an xCELLigence assay with or without re-expression of SPRY1 (DOX, 1 μg/ml). Graphs depict
the growth curves of the cells cultured in the absence or presence of doxycycline during 120 h and they show one representative experimentout of three independent experiments performed. Re-expression of SPRY1 produces a significant inhibition of cell proliferation. Slight artifacts
in the graphs at 72 h are a consequence of media change and subsequent readjustment of the conditions in the xCELLigence device and donot affect the final result. (c) A673/TR/SPRY1, SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were plated in octuplicates and cultured in thepresence or absence of doxycycline (DOX, 1 μg/ml) for 72 h in 10% tetracycline-free FBS-supplemented media (standard culture conditions).
Cell proliferation was assayed by bromodeoxyuridine (BrdU) incorporation into DNA. Graphs depict the percentage of cell proliferationof doxycycline-treated cells (expressing SPRY1) versus control. Figure depicts one representative experiment (mean ± s.d.) out of three
independent experiments performed (***P o0.005). (d) Cells were plated and cultured as described in c, but kept in 1% FBS-supplemented
media (low-serum conditions). Cell proliferation is significantly inhibited in doxycycline-treated cells (expressing SPRY1) versus control. Figure
depicts one representative experiment (mean ± s.d.) out of three independent experiments performed (***Po0.005).
First, we studied the effect of SPRY1 induction on cell proliferation
Finally, the three Ewing cell lines harboring the SPRY1 construct
. Induction of SPRY1 in these Ewing sarcoma cell lines
were cultured in the presence or absence of doxycycline and
on doxycycline stimulation significantly reduced their proliferation.
assayed for cell cycle in non-synchronized cells by flow cytometry.
This was observed using real-time monitoring of cell number
As shown in Supplementary Figure S3, the impairment in cell
(xCELLigence instrument, ACEA Biosciences, San Diego, CA, USA)
proliferation in SPRY1-re-expressing cells seems partially due to a
and by bromodeoxyuridine incorporation assays
cell cycle arrest in the G1 phase, although these results were not
). Notably, no effect on cell proliferation was
observed in cells carrying the empty vector, both in the absence and
Taken together, these results provide evidence that SPRY1
in the presence of doxycycline (data not shown). Cell proliferation
induction impairs cell proliferation as well as clonogenic and
inhibition was observed in standard culture media supplemented
anchorage-independent growth of Ewing sarcoma cell lines.
with 10% fetal bovine serum (FBS) ) and in low-serum (1% FBS) conditions as well Induction of SPRY1 in
SPRY1 induction impairs migration of Ewing sarcoma cells
cells grown in low-serum conditions exhibited an even stronger
We next analyzed the effect of SPRY1 induction in Ewing sarcoma
reduction of cell proliferation (probably suggesting that in
cells on cell migration. As shown in SPRY1 induction
conditions where there is a diminished availability of growth factors,
reduced the ability of A673, SKES and SKNMC Ewing sarcoma
such as in the tumor microenvironment, SPRY1 is able to markedly
cells to close an artificial wound produced in a confluent
impair cell proliferation.
cell monolayer (in vitro wound-healing assay). In addition, SPRY1
Similarly, induction of SPRY1 expression reduced clonogenic
re-expression significantly impaired migration of Ewing sarcoma
growth of the three Ewing sarcoma cell lines plated at very low
cells through a porous membrane (transwell assay) No
density in medium supplemented with 5% FBS, whereas cells
differences in the migratory properties were observed in cells
carrying the empty vector remained unaffected on doxycycline
carrying the empty vector (data not shown).
treatment and Supplementary Figure 2A). When cellswere tested for anchorage-independent growth in soft agar,
SPRY1 repression is necessary for ERK activation and proliferation
no significant differences were observed in the number of
in Ewing sarcoma cells
colonies formed, whereas there was a significant difference in
SPRY1 has been described to inhibit MAPK/ERK pathway, which
the size of the individual colonies No significant
is one of the most relevant proliferative pathways in cancer. For
differences in anchorage-independent growth were observed in
that reason we investigated the effect of SPRY1 induction on ERK
cells transfected with the empty vector when treated accord-
activation mediated by serum. As shown in SPRY1
ingly (Supplementary Figure 2B).
induction reduced the levels of phospho-ERK both in low (1%) and
2016 Macmillan Publishers Limited
Oncogene (2016) 1 – 11
SPRY1 is a tumor suppressor in Ewing sarcoma
F Cidre-Aranaz et al
Figure 3. SPRY1 re-expression impairs Ewing sarcoma cell clonogenicity, anchorage-independent growth, migration and invasion of Ewingsarcoma cells. (a) A673/TR/SPRY1, SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were platted in triplicates at low densities and treated with
or without doxycycline (DOX, 1 μg/ml) for 9 days. Colony formation was measured by crystal violet staining. Pictures show representative wells
of one out of three independent experiments. Graphs depict a quantification of absorbance measured after cell de-staining (one
representative experiment out of three performed) (mean ± s.d.). Clonogenic growth is significantly impaired in all three cell lines on SPRY1
re-expression (*P o0.05, **Po0.01 and ***Po0.005). (b) A673/TR/SPRY1, SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were platted in triplicatein soft agar and cultured in the presence or absence of doxycycline (DOX, 1 μg/ml) during 25 days and subsequently stained with crystal
violet. Pictures show representative images of sphere formation taken at the end of the experiment. Graphs depict the mean area per particleafter 25 days (mean ± s.d.). SPRY1 re-expression inhibits sphere formation in all three cell lines (3 independent experiments) (*Po0.05,
**P o0.01 and ***Po0.005). (c) A673/TR/SPRY1, SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were platted in triplicates and treated with
or without doxycycline (DOX, 1 μg/ml) for 72 h. A ‘wound gap' was created by scratching the cell monolayer using a micropipette tip. Pictures
depict the healing of the gap as a consequence of cell migration at the beginning, middle and end of the experiments. Relative woundclosure for each cell line at the end of the experiment is stated in percentages. Images show a representative experiment out of threeperformed. (d) A673/TR/SPRY1, SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were incubated in the absence or presence of doxycycline (DOX,
1 μg/ml) during 48 h, to induce the expression of SPRY1 protein. Afterwards, cells were starved for another 24 h. Next, they were placed in the
upper compartment of a transwell and allowed to migrate through the membrane in response to serum. Migrating cells were quantified
by crystal violet staining. Figure shows mean ± s.d. of two experiments performed in triplicate. Data are shown as arbitrary units of absorbance
(abs) (*P o0.05 and ***Po0.005).
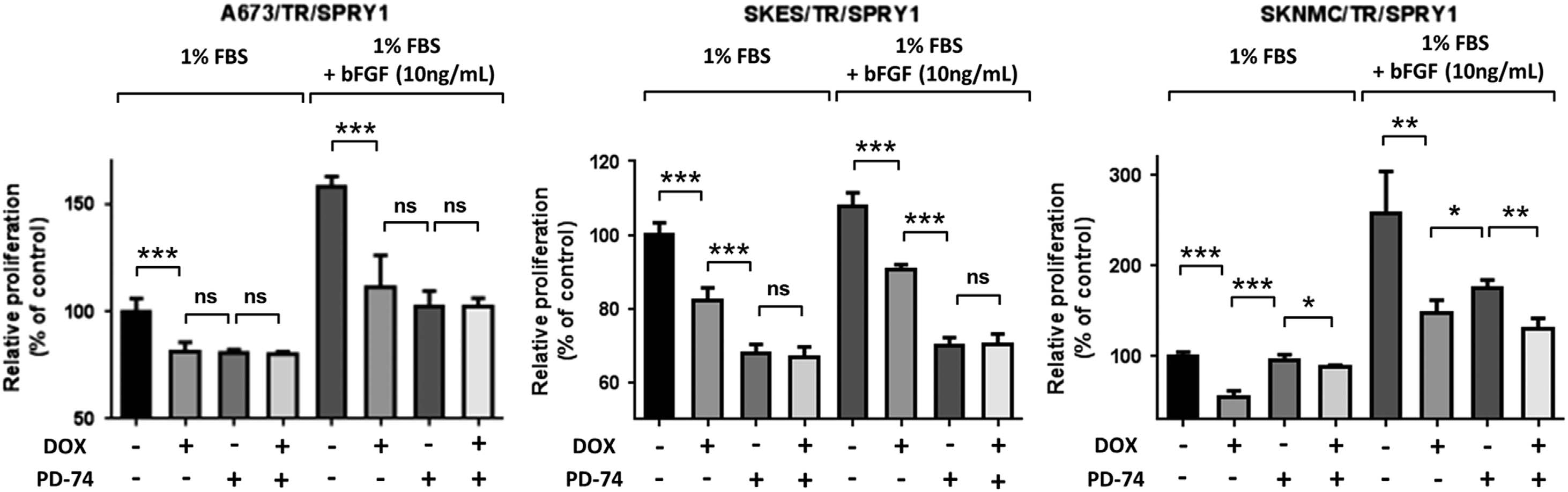
standard (10%) serum conditions. Next, we explored the effect of
Consistently, FGFR inhibition through any of the four FGF
SPRY1 induction on the ERK activation mediated by basic FGF
inhibitors severely impairs clonogenic growth of A673, SKNMC
(bFGF), an established and potent RAS-activating growth factor.
and POE Ewing sarcoma cell lines
Using bFGF stimulation we observed a similar effect on ERK
As POE cells exhibited high sensitivity toward this FGFR
activation in the three Ewing cell lines.
inhibiton compared with the other cells tested (Supplementary
Collectively, these results indicate that EWS-FLI1-mediated SPRY1
Table 2), we chose this cell line to perform in vivo experiments to
repression in Ewing sarcoma cells contributes to the activation of
test whether PD-74 has an antitumoral effect in a xenograft model
MAPK/ERK pathway and thus to the malignant features observed.
in mice. As shown in PD-74 treatment significantlyinhibited tumor growth (P = 0.004) of Ewing sarcoma xenografts.
These tumors showed an 50% decrease in the number of
FGFR inhibitors mimic the effects of SPRY1 re-expression.
mitoses (P = 0.001) along with a 40% increase in the number
As SPRY1 proved to be capable of inhibiting ERK phosphorylation,
of apoptotic cells per high-power field (P = 0.001) when comparing
especially when the FGF pathway was activated, we assessed
vehicle versus PD-74 treatment. Moreover, Ki-67 staining for
the effect of four FGFR inhibitors (PD-173074 [PD-74], PD-166866
proliferation showed a significant reduction in the number
[PD-66], SU5402 [SU54] and NVP-BGJ398 [BG-98]) on Ewing
of Ki67-positive cells in the tumor samples treated with PD-74
sarcoma cells (A673, SKNMC, SKES, RDES and POE), in order
to test whether FGFR inhibition can mimic SPRY1 effect on Ewing
To confirm whether PD-74 had an antitumoral effect in other
cell lines. Complementary to our previous finding that bFGF
Ewing sarcoma cell lines we performed an in vivo experiment
can induce proliferation in Ewing sarcoma cell lines (that is, A673,
using SKES cells, which presented less sensitivity to it in vitro
SKNMC and POE we observed that FGFR inhibition
. As shown in Supplementary Figure S4A, PD-74 had
reduces proliferation of these Ewing sarcoma cells
a dose-dependent effect on SKES xenograft growth in mice,
whereas it did not affect normal cells such as fibroblasts (IMR90).
with 20 mg/kg being the most effective dose (P = 0.005). Again,
Oncogene (2016) 1 – 11
2016 Macmillan Publishers Limited

SPRY1 is a tumor suppressor in Ewing sarcomaF Cidre-Aranaz et al
Figure 4. SPRY1 inhibits MAPK pathway in Ewing sarcoma cells by inhibiting ERK phosphorylation induced by bFGF or serum. A673/TR/SPRY1,SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were incubated in the absence or in the presence of doxycycline (DOX, 1 μg/ml) during 48 h, to
induce the expression of SPRY1 protein. Afterwards, cells were starved for an extra 24 h (1% FBS) and finally stimulated for 15 min with 10%
FBS or bFGF (bFGF, basic fibroblast growth factor) (10 ng/ml) where indicated. SPRY1, phospho-ERK (pERK), ERK and EWS-FLI1 proteins were
detected by specific antibodies. Anti-tubulin was used as a control for loading and transferring. SPRY1 re-expression is capable of inhibiting
ERK phosphorylation induced by bFGF or serum in all three cell lines. Graphs depict densitometries corresponding to the western blottingbands showing pERK/total ERK ratios in percentage versus cells cultured in the absence of doxycycline (control). The figure shows one
representative experiment out of three performed.
we observed a significant reduction of the number of mitoses
of SPRY1 in situ could be associated with clinical outcome in Ewing
(P = 0.01) and a significant increase in the number of apoptotic
sarcoma patients.
cells per high-power field (P o 0.001) on treatment with PD-74
First, SPRY1 mRNA levels were examined in a cohort of 117
(20 mg/kg) (Supplementary Figure S4B). Similarly, we detected
Ewing sarcoma samples studied with gene expression micro-
significantly less Ki-67 positive cells on treatment of with PD-74
arrays and compared them with published microarray data
(P o 0.01) (Supplementary Figure S4B).
sets comprising 24 different solid tumor types.This analysis
We next explored the combined effect of FGFR inhibition and
revealed that Ewing sarcoma range among the ones with the
SPRY1 re-expression. SPRY1 was re-expressed in the three Ewing
lowest SPRY1 expression although in in situ tumors
sarcoma cell lines and they were concomitantly treated with either
there was more heterogeneity in the SPRY1 mRNA levels as
bFGF or PD-74 alone or a combination of both
compared with Ewing sarcoma cells in culture (
In analogy to the results presented in SPRY1 significantly
Moreover, there was statistically less SPRY1 expression in Ewing
inhibited cell proliferation induced by bFGF in the three cell lines
sarcoma cell lines as compared with primary tumors. In fact, all
studied. Moreover, the effect of SPRY1 re-expression and PD-74
cell lines except for one exhibited less SPRY1 expression than
on cell proliferation was similar in A673 and SKNMC cells
the median sample of primary tumors. In contrast, there was no
Furthermore, when the three cell lines were treated
statistical difference in LOX, NR0B1 and CD99 expression in cell
with other FGF inhibitors (BG98, PD-66 and SU54), two of them
lines when compared with primary tumors
(BG98 and PD-66) were able to significantly further reduce the
Next, we analyzed the correlation between SPRY1 levels in
proliferation beyond the effect of SPRY1 alone (Supplementary
primary tumors and clinical outcome in a cohort of 162 Ewing
Figure S5). However, when SPRY1 was re-expressed concomitantly
sarcoma patientsThe median expression value of SPRY1 was
with any of the three new inhibitors tested, none of them
used as a cutoff to define moderate and low SPRY1 expression
produced a further impairment in proliferation on any of the cells
levels. Using this cutoff, moderate SPRY1 expression levels
tested, which is in agreement with what was previously observed
were significantly associated with a better overall survival
with PD-74 (Supplementary Figure S5).
(5-year overall survival 0.70 vs 0.38, P = 0.002; log-rank test)and event-free survival (5-year event-free survival 0.72 vs 0.45,P = 0.0015; log-rank test) (Interestingly,
SPRY1 expression positively correlates with improved overall
low SPRY1 levels were associated with a higher risk for
survival of Ewing sarcoma patients
the presence of metastasis at diagnosis (P = 0.002, Fisher's
Our results indicate that SPRY1 repression leads to a constitutive
exact test) Collectively, these results strongly
activation of MAPK/ERK pathway in response to external stimuli
support a relationship between the levels of SPRY1 and Ewing
such as bFGF. Thus, we wondered whether the expression levels
2016 Macmillan Publishers Limited
Oncogene (2016) 1 – 11
SPRY1 is a tumor suppressor in Ewing sarcoma
F Cidre-Aranaz et al
Figure 5. FGFR inhibitors block Ewing sarcoma cell line proliferation. (a) Four FGFR inhibitors, namely PD-173074 (PD-74), PD-166866 (PD-66),
SU5402 (SU54) and NVP-BGJ398 (BG-98), inhibit A673, SKNMC, POE, RDES and SKES Ewing cell growth in vitro in a dose-dependent manner,whereas normal cells (IMR90 fibroblasts) remained unaffected. PD-74 proved to be most effective in four out of five Ewing sarcoma cell lines
tested. Cells were grown in 10% FBS conditions and cell proliferation was measured after 72 h using a Resazurin assay. (b) PD-173074 (PD-74),
PD-166866 (PD-66), SU5402 (SU54) and NVP-BGJ398 (BG-98) impair A673, SKNMC and POE Ewing sarcoma cells clonogenic growth in vitrowhen cells are grown at 5% FBS for 10–12 days. (c) C.B17/SCID mice were injected with POE cells and randomly split in groups. Each group
was treated intraperitoneally once a day with PD-74 or placebo. The figure shows the evolution of tumor volume (mean ± s.e.m. of six to eight
animals per group) versus time. PD-74 treatment significantly inhibits tumor growth (P = 0.004) of Ewing sarcoma xenografts.
(d) Immunohistochemistry images of tumors obtained in the in vivo experiments. Tissue sections were stained with Ki-67 to detectproliferation and cleaved caspase 3 to detect apoptosis. The graphs show how PD-74 treatment reduces the number of mitoses (P = 0.001)
and increases the number of apoptotic cells per field (P = 0.001). Ki-67 staining and graph show a reduction in the number of Ki67-positive
(++ or +) cells when treated with PD-74 (P o0.01).
SPRY1 promoter directly (Supplementary Figure S6). Interestingly,
EWS-ETS fusion proteins have a central role in the pathogenesis
on EWS-FLI1 knockdown there is an increase of H3K27ac marks
of Ewing sarcoma by regulating the expression of other key
located at the putative SPRY1 promoter comprising SPRY1 exon 1
factors. In this sense, the identification of these regulated genes
and intron 1 (Supplementary Figure S6). This suggests an epigenetic
may help characterize the pathways involved in Ewing sarcoma
mechanism of SPRY1 regulation involving histone modifica-
pathogenesis and aggressiveness, and to therefore open new
tions, instead of a direct binding of EWS-FLI1 to the SPRY1
opportunities for targeted
promoter. Moreover, there were no significant differences in
In this study, we showed that SPRY1, a member of the Sprouty
the percentage of SPRY1 CpG islands' methylation on modula-
family of proteins, is repressed by EWS-FLI1 and is not expressed
tion of EWS-FLI1 expression levels.Accordingly, we propose
in established Ewing sarcoma cell lines. The exact mechanism
that the actual mechanism underlying SPRY1 regulation in Ewing
through which EWS-FLI1 regulates SPRY1 is still unknown. However,
sarcoma may be different from the one operating in other
analysis of two independent chromatin immunoprecipitation
tumors where SPRY1 downregulation is associated with promo-
sequencing studiesindicates that EWS-FLI1 does not bind to
ter methylation.
Oncogene (2016) 1 – 11
2016 Macmillan Publishers Limited

SPRY1 is a tumor suppressor in Ewing sarcomaF Cidre-Aranaz et al
Figure 6. bFGF induces proliferation of Ewing sarcoma cells, which can be antagonized by FGFR inhibition. A673/TR/SPRY1, SKES/TR/SPRY1and SKNMC/TR/SPRY1 cells were incubated in the absence or in the presence of doxycycline (DOX, 1 μg/ml), to induce the expression of
SPRY1 protein, and were concomitantly cultured with 1% FBS, bFGF (10 ng/ml), PD-173074 (PD-74, 5 μM) or a combination of bFGF and PD-74
where indicated. Cell proliferation was measured after 72 h using the Resazurin assay. Graphs depict one independent experiment (mean ± s.d.)
out of three performed. SPRY1 re-expression and PD-74 inhibit cell proliferation-induced serum or bFGF treatment (*Po0.05 and **Po0.005, ns,
Figure 7. SPRY1 expression is positively correlated with improved overall survival of Ewing sarcoma patients. (a) SPRY1 mRNA expressionlevels in 24 different solid tumor entities as determined by Affymetrix HG-U133plus2.0 DNA microarrays. Data were retrieved from the GeneExpression Omnibus (GEO) or the European bioinformatics Institute (EBI) and simultaneously normalized by RMA using brain array CDF files
(v17, ENTREZG) as previously described.Data are represented as medians with boxes representing the interquartile range. Whiskers indicatethe 10th and 90th percentile of the data. The number of analyzed samples is given in parentheses. Ewing sarcoma tumors are shown in gray.
ATRT, atypical teratoid rhabdoid tumor; Ca-, carcinoma; GIST, gastrointestinal stromal tumor; NSCLC, non-small cell lung carcinoma; RMS,rhabdomyosarcoma. (b) Relative expression of SPRY1 as compared with other EWS-FLI1 target genes (LOX and NR0B1) and CD99 in 15individual Ewing sarcoma cell lines versus 117 primary Ewing sarcoma samples (all Affymetrix HG-U133Plus2.0 microarrays). Data wereretrieved from the GEO (accession codes: GSE8596, GSE36133, GSE70826 and GSE34620) and simultaneously normalized by RMA usingbrainarray CDF files (v17, ENTREZG) as previously described.Unpaired two-tailed Student's T-test. (c) Kaplan–Meier survival estimates (overall
survival) in the Ewing sarcoma patient cohort. Patients were classified as being either SPRY1 low or moderate (cutoff: median SPRY1
expression; P = 0.002, log-rank test). (d) Graph depicts the relapse-free survival probability versus SPRY1 level of expression (low or moderate,
cutoff: median SPRY1 expression). SPRY1 expression positively correlates with improved relapse-free probability (P = 0.0015, log rank test).
(e) Graph shows the percentage of cases with metastasis at diagnosis versus SPRY1 level of expression (low or moderate, cutoff: median SPRY1expression). Moderate SPRY1 expression correlates with lower risk of metastasis at diagnosis (P = 0.002, Fisher's exact test).
2016 Macmillan Publishers Limited
Oncogene (2016) 1 – 11
SPRY1 is a tumor suppressor in Ewing sarcoma
F Cidre-Aranaz et al
As SPRY1 has been shown to be a potent negative regulator of
however, it can be hypothesized that SPRY1 levels remain variable
the RAS/MAPK/ERK signaling we hypothesized that
in tumors, and that the harsh conditions of in vitro cell culture
SPRY1 may act as a tumor suppressor gene in Ewing sarcoma.
favor the growth of cells with lower SPRY1 levels during
In support of this notion, induction of SPRY1 in three independent
establishment of Ewing sarcoma cell lines. In fact, established
Ewing sarcoma cell lines significantly impaired cell prolifera-
Ewing sarcoma cell lines harbor a much higher rate of STAG2, TP53
tion and migration. This is consistent with a tumor supressor
and CDKN1A mutations than that observed in primary tumors
function of SPRY1 in Ewing sarcoma and in agreement with
suggesting that cells derived from more aggressive
previous reports showing that SPRY1 overexpression impairs cell
tumors are favored in
growth, proliferation, migration and invasion of a variety of cancer
Ewing sarcoma is a very aggressive pediatric malignancy
cell lines including ovarian carcinoma, breast cancer, lung
in which primary metastasis is the most unfavorable risk factor,
adenocarcinoma, colon carcinoma or osteosarcoma.
very often leading to fatal outcome despite highly intense and
Our results also demonstrate that SPRY1 downregulation
toxic Here we show that low SPRY1 expression levels
is necessary for bFGF-mediated proliferation and activation of
correlate with a significantly worse overall and event-free survival
RAS/MAPK/ERK pathways in Ewing sarcoma cells. Thus, SPRY1
in a large cohort of Ewing sarcoma patients. More interestingly,
re-expression in the three Ewing sarcoma cell lines used in this
primary tumors displaying low levels of SPRY1 were more
study impaired cell proliferation and ERK phosphorylation induced
frequently observed in patients harboring metastasis at diagnosis.
by bFGF. bFGF is known to mediate proliferation, migration and
This is compatible with a more aggressive behavior of SPRY1-low
differentiation in various cellular and FGF-regulated
tumors and in agreement with the results observed in the in vitro
pathways have a preponderant role in cancer (reviewed in Touat
experiments. We speculate that tumors expressing low levels
et al.Notably, an important role for FGF-dependent pathways
of SPRY1 would present a higher response to external growth
in Ewing sarcoma pathogenesis is emerging. We have recently
factor stimulation and thus exhibit higher rates of proliferation
reported that bFGF increases proliferation of Ewing sarcoma cells
and migration, making them more aggressive. This may have
in vitro, and that EGR2, which is a downstream component of the
a potential clinical application, as SPRY1 has been recently
FGF pathway, is an EWS-FLI1-induced target gene.Other studies
proposed as a possible tissue biomarker to differentiate aggressive
have demonstrated that bFGF regulates motility and invasion of
from indolent prostate carcinomas.
Ewing sarcoma cells in the bone microenvironmentIn agreement,
In summary, our data provide evidence that EWS-FLI1-mediated
Agelopoulos et al.recently showed that constitutive knockdown of
SPRY1 downregulation is an important mechanism in Ewing
FGFR1 abolishes engraftment of Ewing sarcoma xenografts in mice.
sarcoma pathogenesis. Moreover, our results strongly suggest that
Interestingly, over 75% of Ewing sarcoma biopsy samples present
bFGF-mediated stimulation of cell proliferation could be more
moderate-to-high levels of FGFR1 palthough
important than initially acknowledged in Ewing sarcoma, and that
activating FGFR1 mutations are extremely rare in this
FGFR inhibitors may constitute promising drugs for treatment
In light of these facts and our new results, we propose that
of Ewing sarcoma patients.
constitutive activation of FGFRs and downstream pathwaysare key contributors to the pathogenesis of Ewing sarcoma, andthat EWS-FLI1-mediated suppression of the negative-feedback
MATERIALS AND METHODS
regulator SPRY1 constitutes a major mechanism for sustained
FGFR phosphorylation and thus unrestrained FGF-induced signal
A673/TR/shEF cells, which have been described elsewhere,were cultured
transduction and tumor progression. In synopsis, our results
in Dulbecco's modified Eagle's medium supplemented with 10%
support that SPRY1 downregulation is pre-requisite for enhanced
proliferation and migration of Ewing sarcoma cells induced
100 μg/ml zeocin and 3 μg/ml blasticidin. Induction of
by either EWS-FLI1 itself, external growth factor stimulation or
a small hairpin RNA against EWS-FLI1 was started by the addition
a combination of both as part of an autocrine loop.
of 1 μg/ml doxycycline (Sigma, St Louis, MO, USA). Ewing sarcoma cell lines
The importance of this pathway in Ewing sarcoma pathogenesis
A4573, TC-71, RD-ES, POE and TTC-466, and the normal fibroblast cell
is additionally illustrated by FGFR-inhibition-mediated impairment
line IMR90 were maintained in RPMI 1640 medium, SK-PN-DW and SKNMCwild-type cells were maintained in Iscove's modified Dulbecco's medium,
of cell proliferation and clonogenic growth of Ewing sarcoma cells
and wild-type A673 and SKES cells were maintained in Dulbecco's
in vitro. Interestingly, the search for more efficient and specific
modified Eagle's medium. All media were supplemented, if not other-
FGFR inhibitors is an active field in the pharmaceutical industry,
wise stated, with 10% FBS, 2 mM L-glutamine (Invitrogen, Paisley, UK)
as FGF signaling pathways is one of the most commonly mutated
and 1% penicillin and streptomycin. All cells were routinely tested
systems in cancer.In this regard, the Ewing sarcoma research
for mycoplasma contamination (Mycoalert mycoplasma detection kit,
community can take advantage of the development of these new
Lonza #LT07-318, Basel, Switzerland) and were authenticated by short
drugs, some of which are being tested in clinical trials with
tandem repeats profiling at the Genomic Facility at Biomedical Research
promising results, particularly in tumors harboring aberrant FGFR
Institute, CSIC, Madrid, Spain).
signaling (reviewed in Touat et al.).
FGFR signaling can be aberrantly activated in Ewing sarcoma
Establishment of Ewing sarcoma cell lines stably expressing
either through downregulation of SPRY1 (as observed in most
doxycycline-inducible SPRY1 cDNA
cases), through overexpression of FGFRs (as observed in subset
The complete coding region of SPRY1 was reverse transcription–PCR
of pa), or very rarely through somatic mutaFor
amplified from A673/TR/shEF cells stimulated with doxycycline using primers
that reason, we anticipate that Ewing sarcoma patients may benefit
5′-GCGGTCGACGAGATCACTACACATGGATCC-3′ (forward) and 5′-CGGCGGCC
from targeted drugs directed against FGFRs or its downstream
GCTCATCATCATGATGGTTTACCCTGACC-3′ (reverse). The amplified fragments
targets. In support of this notion, Agelopoulus et reported on
were digested with SalI and NotI, cloned into the pENTR2B plasmid
a single patient affected by relapsed Ewing sarcoma, who was
(Invitrogen) and transferred by recombination to the lentiviral doxycycline-
treated with an FGFR-tyrosine kinase inhibitor (ponatinib), which led
inducible plasmid pLenti4-TO-V5-DEST (Invitrogen). Next, A673/TR, SKES/TR
to a reduction in 18-FDG-PET activity and thus glucose uptake by
and SKNMC/TR Ewing sarcoma cells expressing the tetracycline repressorconstitutively were infected with lentiviruses containing the SPRY1 cDNA.
Control cells were infected with empty lentiviral vector. Stable clones were
Interestingly, although SPRY1 was undetectable in established
selected with zeocin (100 μg/ml). Induction of SPRY1 was assayed by western
Ewing sarcoma cell lines, its levels in primary tumors were variable.
blottings on doxycycline (1 μg/ml) stimulation. Clones displaying the highest
Currently, the reasons for the differences between SPRY1 levels
levels of protein expression on doxycycline stimulation were chosen for
in established cell lines and tumors in situ are still unknown;
additional studies.
Oncogene (2016) 1 – 11
2016 Macmillan Publishers Limited
SPRY1 is a tumor suppressor in Ewing sarcomaF Cidre-Aranaz et al
Reverse transcription–quantitative PCR
Reverse transcription–quantitative PCR conditions, primer and TaqMan
Cells were pre-treated with doxycycline (1 μg/ml) for 24 h to induce the
probe sequences specific for EWS-FLI1, LOX, NR0B1(DAX1) and TBP were
expression of SPRY1 protein. Next, they were starved (0.5% FBS) for
described TaqMan probes for SPRY1, 2, 3 and 4 were
another additional 24 h in the same doxycycline conditions. Then, 3 × 105
purchased to Life Technologies (San Diego, CA, USA). Reactions were run
pretreated cells were re-suspended in 2 ml of medium containing 0.5%
on a RotorGene 6000 (Corbett Research, Sydney, NSW, Australia) and
tetracycline-free FBS and placed in the upper chamber of transwells
relative expression was calculated as previously described
(8.0 μm pore size) (Corning Costar, Cambridge, MA, USA) following theprocedure described
Western blot analysis and antibodies
Procedure was described elsewhere.Primary antibodies were purchased to
Cells were plated by triplicate (5 × 105 cells per 60 mm dishes) in soft agar
the following companies: anti-FLI1 polyclonal antibody from NeoMarkers
and cultured in the presence or absence of doxycycline during 25 days.
(#RB-9295-P) (Fremont, CA, USA), anti-SPRY1 monoclonal antibody from
Fresh culture medium was added to plates every 2–3 days. At the end of the
Santa Cruz Biotechnology (#100861) (Dallas, TX, USA), Tubulin monoclonal
experiment, three random fields for each plate were photographed. The
antibody from Sigma Aldrich (#T9026) (St Louis, MO, USA), and anti-Phospho-
number of colonies per field and its respective area were calculated using
p44/42 (pERK, #9106) and anti-p44/42 (totalERK, #9102) were from Cell
NIH ImageJ software (National Institute of Health, Bethesda, MD, USA).
Signaling (Danvers, MA, USA). Anti-mouse (#2055) and anti-rabbit IgG (#2054)horseradish peroxidase-conjugated secondary antibodies were purchased
from Santa Cruz Biotechnology.
A673/TR/SPRY1, SKES/TR/SPRY1 and SKNMC/TR/SPRY1 cells were platedin triplicates at 0.5 × 103, 1 × 103 and 2 × 103 cells per well, respectively,
Bromodeoxyuridine proliferation assay
in a 24-well plate. They were subsequently treated with or without
Cells were plated in octaplicates (1 × 103 cells per well in 96 multi-well
doxycycline (1 μg/ml) and maintained for 9 days in culture media
plates) and cultured in the presence or absence of doxycycline (1 μg/ml)
supplemented with 5% tetracycline-free FBS. Media was changed every
for 72 h in 10% or 1% tetracycline-free FBS (Clontech). Thereafter,
3–4 days and doxycycline treatment was continued. Finally, colonieswere fixed, stained with crystal violet and photographed. Cells were
bromodeoxyuridine chemiluminescent assay (Roche, Basel, Switzerland)
de-stained using 50% ethanol 0.1 M sodium citrate pH 4.2. Absorbance was
was performed according to manufacturer's instructions. Chemiluminescence
quantified at 560 nm using an Infinite M200 (Tecan) microplate reader.
was measured using an Infinite M200 (Tecan, Mannerdorf, Switzerland)microplate reader.
Tumor xenografts in micePOE and SKES cells were resuspended in PBS/matrigel (BD Biosciences,
Resazurin proliferation assay
Le Pont de Claix Cedex, France) (1:1) and injected (8 × 106/200 μl)
Cells were plated in octaplicates (2.5 × 103 cells per well in 96 multi-well
subcutaneously in the flanks of 6-week old C.B17/SCID male and female
plates) and concomitantly cultured in the presence or absence
mice (Charles River Laboratories, Lyon, France). When tumor volume
of doxycycline (1 μg/ml) and stimuli (1% or 10% tetracycline-free FBS
reached 150 mm3 (calculated with the formula length × width × depth ×
or 10 ng/ml bFGF) for 72 h. For bFGF-inhibitor testing, cells were grown at
0.5432), mice were injected intraperitoneally once a day with the indicated
10% FBS for 72 h in the presence of PD-173074 (PD-74) (Selleckchem,
dose of PD-173074 (5, 10 or 20 mg/kg) dissolved in 10% dimethyl
Houston, TX, USA), PD-166866 (PD-66) (#PZ0114, Sigma Aldrich), SU5402
sulfoxide–90% Corn Oil (Sigma) or placebo in the control group. Tumor
(SU54) (#S7667, Selleckchem) or NVP-BGJ398 (BG-98) (#S2183, Selleck-
growth was monitored with a caliper and mice were killed when tumors
chem). Thereafter, Resazurin (#R7017, Sigma Aldrich) was added to the
reached a volume of 1500 mm3. Experiments were carried out in
media at 0.15 μg/ml and incubated for 2 h at 37 °C. Fluorescence was
accordance with recommendations of the European Community (86/609/EEC), the French Competent Authority, the UKCCCR guidelines (guidelines
recorded using a 560-nm excitation/590-nm emission filter set in an
for the welfare and use of animals in cancer research), the Ethics
Infinite M200 microplate reader (Tecan).
Committee at ISCIII (CBA #64_2015-v2) and the Spanish CompetentAuthority (PROEX 009/16).
xCELLigence proliferation assayCell proliferation was assayed in real time with a bioelectric xCELLigence
Histology and immunohistochemistry
instrument (Roche/ACEA Biosciences). In each well, 3–4 × 103 Ewing
Immunohistochemistry analyses were done on formalin-fixed, paraffin-
sarcoma cells were seeded in 200 μl media containing 10% tetracycline-
embedded xenograft tumors. All tissue samples were collected at
free FBS and treated with doxycycline (1 μg/ml) or vehicle (triplicate wells/
the Institute of Pathology of the LMU Munich for immediate immunohis-
group). Cellular impedance was measured periodically and media with
tochemistry staining, for which 4-μm sections were cut. Antigen retrieval was
or without doxycycline were changed once after 72 h.
carried out by microwave treatment in Dako target retrieval solution (S2369).
The following primary antibodies were used: polyclonal rabbit anti-cleaved-caspase-3 (1:100 at room temperature for 60 min; #9661, Cell Signaling) or
Flow cytometry analysis of cell cycle
monoclonal rabbit anti-Ki67 (1:200 at room temperature for 60 min;
Cells were treated with doxycycline (1 μg/ml) for 72 h to induce the
#275R-15 clone SP6, Cell Marque, Rocklin, CA, USA). The ImmPRESS Reagent
expression of SPRY1 and fixed with 70% ethanol for 24 h at 4 °C. Next, they
Kit anti-rabbit IgG (MP-7401, Vector Laboratories, Burlingame, CA, USA) was
were stained with a solution of 0.005% (w/v) of propidium iodide and
used for antigen detection. Sections were counterstained with hematoxylin
RNAase A as recommended by the manufacturer (BD Biosciences, San José,
Gill's Formula (H-3401, Vector Laboratories). The average number of positive
CA, USA) and were incubated at 37 °C for 30 min. They were then analyzed
cells was determined by analysis of 10 high-power fields (×40 magnification)
in a MACS Quant Analyzer flow cytometer (Miltenyi Biotec, Cologne,
for each xenograft tumor. Statistical differences between groups were
calculated with an unpaired tow-tailed Student's T-test.
Wound-healing assay
A total of 162 Ewing sarcoma patients with available clinical data and
Cells were plated in triplicates (2–4 × 104 cells per well in 24 multi-well
tumor samples were used in this study. This cohort consists of 117 Ewing
plates) and were incubated with or without doxycycline (1 μg/ml) for 72 h
patients for which gene expression profiles in primary tumors were
before the assay. At the end of this period, a ‘wound gap' in the cell
analyzed with HG-U133 plus2.0 microarrays (Affymetrix, Santa Clara, CA,
monolayer was created using a micropipette tip. The healing of the gap
USA) (Gene Expression Omnibus accession number: GSE34620) and 45
by cell migrating was monitored by photographing the progress every
patients whose gene expression profiles were studied with Uniset Human
6–12 h until wound closure. Quantification of relative cell migration is
20 K I microarrays (Codelink Amersham Bioscience, Piscataway, NJ, USA).
All patients received a similar protocol treatment.
2016 Macmillan Publishers Limited
Oncogene (2016) 1 – 11
SPRY1 is a tumor suppressor in Ewing sarcoma
F Cidre-Aranaz et al
Statistical analysis
12 Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling
For a single comparison of two groups, two-tailed Student's t-test was used
of Ewing cells reveal downstream oncogenic pathways and a crucial role
and a normal distribution was assumed. Variances between the groups
for repression of insulin-like growth factor binding protein 3. Mol Cell Biol 2004;
that were compared were similar. For animal studies, the sample size was
24: 7275–7283.
estimated to be six to eight mice considering a signal/noise ratio of 1.6–1.8,
13 Agra N, Cidre F, Garcia-Garcia L, de la Parra J, Alonso J. Lysyl oxidase is
80% power, assuming a 5% significance level and a two-sided test.
downregulated by the EWS/FLI1 oncoprotein and its propeptide domain displays
No investigator blinding was done during the experiment. For in situ
tumor supressor activities in ewing sarcoma cells. PLoS One 2013; 8: e66281.
studies including overall survival and relapse-free survival probabilities,
14 Navarro D, Agra N, Pestana A, Alonso J, Gonzalez-Sancho JM. The EWS/FLI1
log-rank test was used. For proportions, Fisher's exact test was used. For all
oncogenic protein inhibits expression of the Wnt inhibitor DICKKOPF-1 gene and
analyses, the level of significance was set at P = 0.05 and the variance was
similar between groups. All statistical calculations were performed using
31: 394–401.
the GraphPad Prism software version 6.0 (GraphPad Software, San Diego,
15 Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG et al. Repression of the gene
encoding the TGF-beta type II receptor is a major target of the EWS-FLI1oncoprotein. Nat Genet 1999; 23: 222–227.
16 Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N et al. Vertebrate
CONFLICT OF INTEREST
Sprouty genes are induced by FGF signaling and can cause chondrodysplasia
The authors declare no conflict of interest.
when overexpressed. Development 1999; 126: 4465–4475.
17 Guy GR, Wong ES, Yusoff P, Chandramouli S, Lo TL, Lim J et al. Sprouty: how does
the branch manager work? J Cell Sci 2003; 116: 3061–3068.
18 Christofori G. Split personalities: the agonistic antagonist Sprouty. Nat Cell Biol
2003; 5: 377–379.
FC-A, LG-G, JCL, AS, PG-M, SEL-P, SM and JA are supported by Asociación Pablo
19 Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K et al. p53 loss
Ugarte and Miguelañez SA, ASION-La Hucha de Tomás, Fundación La Sonrisa de Alex
promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev
and Instituto de Salud Carlos III (PI12/00816 and Spanish Cancer Network RTICC
2010; 24: 1389–1402.
RD12/0036/0027). TGPG is supported by a grant from ‘Verein zur Förderung von
20 Fritzsche S, Kenzelmann M, Hoffmann MJ, Muller M, Engers R, Grone HJ et al.
Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München
Concomitant down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr
(WiFoMed)', the Daimler and Benz Foundation in cooperation with the Reinhard
Relat Cancer 2006; 13: 839–849.
Frank Foundation, by LMU Munich's Institutional Strategy LMUexcellent within the
21 Lo TL, Yusoff P, Fong CW, Guo K, McCaw BJ, Phillips WA et al. The ras/mitogen-
framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke
activated protein kinase pathway inhibitor and likely tumor suppressor proteins,
Kinder—Bettina-Bräu-Stiftung', the Walter Schulz Foundation, the Fritz Thyssen
sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res 2004; 64:
Foundation (FTH-40.15.0.030MN) and by the German Cancer Aid (DKH-111886 and
DKH-70112257). The ‘Genetics and Biology of Cancers' team (TGPG, DS and OD) is
22 Kwabi-Addo B, Ren C, Ittmann M. DNA methylation and aberrant expression
supported by grants from the Ligue Nationale Contre Le Cancer (Equipe labellisée).
of Sprouty1 in human prostate cancer. Epigenetics 2009; 4: 54–61.
This work was also supported by the European PROVABES, ASSET and EEC FP7 grants.
23 Kwabi-Addo B, Wang J, Erdem H, Vaid A, Castro P, Ayala G et al. The expression
We also thank the following associations for their invaluable support: the Société
of Sprouty1, an inhibitor of fibroblast growth factor signal transduction,
Française des Cancers de l'Enfant, Courir pour Mathieu, Dans les pas du Géant, Olivier
is decreased in human prostate cancer. Cancer Res 2004; 64: 4728–4735.
Chape, Les Bagouzamanon, Enfants et Santé and les Amis de Claire. We thank Dr SNavarro (University of Valencia, Valencia, Spain) and Dr TJ Triche (Children's Hospital
24 Masoumi-Moghaddam S, Amini A, Ehteda A, Wei AQ, Morris DL. The expression
Los Angeles, Los Angeles, USA) for providing us with Ewing sarcoma cell lines A4573
of the Sprouty 1 protein inversely correlates with growth, proliferation, migration
and TTC-466, respectively.
and invasion of ovarian cancer cells. J Ovarian Res 2014; 7: 61.
25 Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer
drug sensitivity. Nature 2012; 483: 603–607.
26 Grunewald TG, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud MM
1 Mackintosh C, Madoz-Gurpide J, Ordonez JL, Osuna D, Herrero-Martin D.
et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2
The molecular pathogenesis of Ewing's sarcoma. Cancer Biol Ther 2010; 9:
via a GGAA microsatellite. Nat Genet 2015; 47: 1073–1078.
27 Willier S, Butt E, Grunewald TG. Lysophosphatidic acid (LPA) signalling in cell
2 Grohar PJ, Helman LJ. Prospects and challenges for the development of new
migration and cancer invasion: a focussed review and analysis of LPA receptor
therapies for Ewing sarcoma. Pharmacol Ther 2013; 137: 216–224.
gene expression on the basis of more than 1700 cancer microarrays. Biol Cell
3 Ladenstein R, Potschger U, Le Deley MC, Whelan J, Paulussen M, Oberlin O et al.
2013; 105: 317–333.
Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING
28 Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H et al. Common
99 trial. J Clin Oncol 2010; 28: 3284–3291.
variants near TARDBP and EGR2 are associated with susceptibility to Ewing
4 Zhu L, McManus MM, Hughes DP. Understanding the biology of bone sarcoma
sarcoma. Nat Genet 2012; 44: 323–327.
from early initiating events through late events in metastasis and disease
29 Cidre-Aranaz F, Alonso J. EWS/FLI1 target genes and therapeutic opportunities
progression. Front Oncol 2013; 3: 230.
in Ewing sarcoma. Front Oncol 2015; 5: 162.
5 Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M et al. Gene fusion
30 Bilke S, Schwentner R, Yang F, Kauer M, Jug G, Walker RL et al. Oncogenic ETS
with an ETS DNA-binding domain caused by chromosome translocation in humantumours. Nature 1992; 359: 162–165.
fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer.
6 Kovar H. Blocking the road, stopping the engine or killing the driver? Advances in
Genome Res 2013; 23: 1797–1809.
targeting EWS/FLI-1 fusion in Ewing sarcoma as novel therapy. Expert Opin Ther
31 Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML et al. EWS-FLI1
Targets 2014; 18: 1315–1328.
utilizes divergent chromatin remodeling mechanisms to directly activate
7 Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I et al.
or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014; 26: 668–681.
32 Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P et al.
tumor growth. Clin Cancer Res 2007; 13: 2429–2440.
Epigenome mapping reveals distinct modes of gene regulation and widespread
8 Garcia-Aragoncillo E, Carrillo J, Lalli E, Agra N, Gomez-Lopez G, Pestana A et al.
enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep 2015;
DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle
10: 1082–1095.
progression in Ewing's tumor cells. Oncogene 2008; 27: 6034–6043.
33 Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA et al. Mechanistic
9 Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR et al. Expression
significance of aberrant methylation in the molecular
profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma.
pathogenesis of human hepatocellular carcinoma. J Clin Invest 2007; 117:
Cancer Cell 2006; 9: 405–416.
10 Surdez D, Benetkiewicz M, Perrin V, Han ZY, Pierron G, Ballet S et al. Targeting
34 Macia A, Gallel P, Vaquero M, Gou-Fabregas M, Santacana M, Maliszewska A et al.
the EWSR1-FLI1 oncogene-induced protein kinase PKC-beta abolishes ewing
Sprouty1 is a candidate tumor-suppressor gene in medullary thyroid carcinoma.
sarcoma growth. Cancer Res 2012; 72: 4494–4503.
Oncogene 2012; 31: 3961–3972.
11 Grunewald TG, Diebold I, Esposito I, Plehm S, Hauer K, Thiel U et al. STEAP1
35 Gross I, Bassit B, Benezra M, Licht JD. Mammalian sprouty proteins inhibit cell
is associated with the invasive and oxidative stress phenotype of Ewing tumors.
growth and differentiation by preventing ras activation. J Biol Chem 2001; 276:
Mol Cancer Res 2012; 10: 52–65.
Oncogene (2016) 1 – 11
2016 Macmillan Publishers Limited
SPRY1 is a tumor suppressor in Ewing sarcomaF Cidre-Aranaz et al
36 Mekkawy AH, Pourgholami MH, Morris DL. Human Sprouty1 suppresses growth,
45 Agelopoulos K, Richter GH, Schmidt E, Dirksen U, von Heyking K, Moser B et al.
migration, and invasion in human breast cancer cells. Tumour Biol 2014; 35:
Deep sequencing in conjunction with expression and functional analyses reveals
activation of FGFR1 in Ewing sarcoma. Clin Cancer Res 2015; 21: 4935–4946.
37 Wiles ET, Lui-Sargent B, Bell R, Lessnick SL. BCL11B is up-regulated by EWS/FLI and
46 Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A et al. Genomic
contributes to the transformed phenotype in Ewing sarcoma. PLoS ONE 2013;
landscape of Ewing sarcoma defines an aggressive subtype with co-association of
STAG2 and TP53 mutations. Cancer Discov 2014; 4: 1342–1353.
38 Liu X, Lan Y, Zhang D, Wang K, Wang Y, Hua ZC. SPRY1 promotes the degradation
47 Kovar H, Jug G, Aryee DN, Zoubek A, Ambros P, Gruber B et al. Among genes
of uPAR and inhibits uPAR-mediated cell adhesion and proliferation. Am J Cancer
involved in the RB dependent cell cycle regulatory cascade, the p16 tumor
Res 2014; 4: 683–697.
suppressor gene is frequently lost in the Ewing family of tumors. Oncogene 1997;
39 Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors
15: 2225–2232.
and signaling. Endocr Relat Cancer 2000; 7: 165–197.
48 Kovar H, Pospisilova S, Jug G, Printz D, Gadner H. Response of Ewing tumor cells
40 Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate
to forced and activated p53 expression. Oncogene 2003; 22: 3193–3204.
development. Endocr Rev 2005; 26: 63–77.
49 Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC et al. Ewing
41 Chalkiadaki G, Nikitovic D, Berdiaki A, Sifaki M, Krasagakis K, Katonis P et al.
sarcoma: current management and future approaches through collaboration.
Fibroblast growth factor-2 modulates melanoma adhesion and migration through
J Clin Oncol 2015; 33: 3036–3046.
a syndecan-4-dependent mechanism. Int J Biochem Cell Biol 2009; 41: 1323–1331.
42 Yamaguchi F, Saya H, Bruner JM, Morrison RS. Differential expression of two
50 Terada N, Shiraishi T, Zeng Y, Aw-Yong KM, Liu Z, Takahashi S et al. Correlation of
fibroblast growth factor-receptor genes is associated with malignant progression
Sprouty1 and Jagged1 with aggressive prostate cancer cells with different sen-
in human astrocytomas. Proc Natl Acad Sci USA 1994; 91: 484–488.
sitivities to androgen deprivation. J Cell Biochem 2014; 115: 1505–1515.
43 Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC. Targeting FGFR signaling
51 Mendiola M, Carrillo J, Garcia E, Lalli E, Hernandez T, de Alava E et al. The orphan
in cancer. Clin Cancer Res 2015; 21: 2684–2694.
nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly
44 Kamura S, Matsumoto Y, Fukushi JI, Fujiwara T, Iida K, Okada Y et al. Basic
expressed in Ewing tumors. Int J Cancer 2006; 118: 1381–1389.
fibroblast growth factor in the bone microenvironment enhances cell motility and
52 Yue PY, Leung EP, Mak NK, Wong RN. A simplified method for quantifying
invasion of Ewing's sarcoma family of tumours by activating the FGFR1-PI3K-Rac1
cell migration/wound healing in 96-well plates. J Biomol Screen 2010; 15:
pathway. Br J Cancer 2010; 103: 370–381.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
2016 Macmillan Publishers Limited
Oncogene (2016) 1 – 11
Source: http://www.asociacionpablougarte.es/pdf/ayudaISCIII/2016_Oncogene.pdf
36V 14.5Ah eZee Flat BatteryCustomized for Grin Tech Specifications and User Guide 35 cm (13.75") Grin Technologies Ltd.20 E 4th AveVancouver, BC, CanadaV5T 1E8 Copyright © 2012 Congratulations on your purchase of this 36V 14.5Ah lithium manganese ebike battery pack. Our hope is that it provides you with a solid 2-3 years of trouble-free power for your vehicle project.
Magnesium Research 2010; 23 (2): 1-13 Magnesium and cardiovascular system Leviev Heart Center, Chaim Sheba Medical Center, Tel Hashomern and the Sackler Facultyof Medicine, Tel Aviv University, Ramat Aviv, IsraelCorrespondence: M.Shechter, MD, MA, FESC, FACC, FAHA, FACN, Director, Clinical Research Unit, Leviev Heart Center, Chaim Sheba Medical Center, 52621 Tel Hashomer, Israel





