Kamagra gibt es auch als Kautabletten, die sich schneller auflösen als normale Pillen. Manche Patienten empfinden das als angenehmer. Wer sich informieren will, findet Hinweise unter kamagra kautabletten.
1471-2180-7-12.fm
Methodology article
A recombinase system facilitates cloning of expression cassettes in
the ciliate Tetrahymena thermophila
Thomas Weide*1, Ulrike Bockau2,3, Angelika Rave2, Lutz Herrmann2,4 and
Marcus WW Hartmann*2
Address: 1Universitaetskliniken Muenster (UKM), Abteilung für Molekulare Nephrologie, Domagkstr. 3a, D-48149 Muenster, Germany, 2Cilian AG, Johann-Krane-Weg 42, D-48149 Muenster, Germany, 3Institut für allgemeine Zoologie und Genetik, Universitaet Muenster, Schloßplatz 5, D-48149 Muenster, Germany and 4Provendis GmbH, Eppinghofer Str. 50, 48468 Muelheim an der Ruhr, Germany
Email: Thomas Weide* -
[email protected]; Ulrike Bockau -
[email protected]; Angelika Rave -
[email protected]; Lutz Herrmann -
[email protected]; Marcus WW Hartmann* -
[email protected]
* Corresponding authors
Published: 1 March 2007
Received: 31 August 2006Accepted: 1 March 2007
BMC Microbiology 2007,
7:12
2007 Weide et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Background: Tetrahymena thermophila is one of the best characterized unicellular eukaryotes and
its genome is sequenced in its entirety. However, the AT-richness of the genome and an unusual
codon usage cause problems in cloning and expression of the ciliate DNA. To overcome these
technical hiatuses we developed a
Cre-dependent recombinase system.
Results: We created novel donor and acceptor vectors that facilitate the transfer of expression
cassettes from the donor into novel acceptor plasmid. Expression vectors were used that encode
the 19 kDa C-terminus of the MSP1 protein of
Plasmodium falciparum and a blasticidin S (bsdR)
resistance gene, respectively. The functional expression of these genes was demonstrated by
western blot analysis with MSP1 specific antibodies and by a blasticidin growing assay.
Conclusion: The
Cre dependent recombinase system in combination with the modular structure
of the donor vectors ease cloning and expression of foreign genes in the ciliate system, providing
a powerful tool for protistology research in future.
Ciliates characteristically possess two nuclei, a somatic
The ciliate
Tetrahymena thermophila has been successfully
macronucleus (MAC) and a germline micronucleus
used as a model system in molecular and cell biology for
(MIC)[tly, the entire genome of the MAC of
T.
decades. Fundamental discoveries such as ribozymes, tel-
thermophila has been characteri]. A shotgun sequenc-
omeric repeats, telomerases or the function of scan RNAs
ing analysis of the MAC revealed that
T. thermophila is 104
were first studied in this eukaryotic microorganism
Mb in length and has approximately 225 MAC chromo-
In addition, cells grow fast to high cell densities in inex-
somes that contain more than 27,000 protein coding
pensive media and simple bioreactor infrastructure and
genes. About 15,000 genes match genes of other organ-
several foreign proteins have been expressed, suggesting
isms. In addition to that the genome analysis also eluci-
that
T. thermophilahas the potential to become an excel-
dated that a huge number of genes are based on gene
lent expression ho].
duplication mechanisms. This is especially true for genesthat play a role in structural complexity, sensing and
(page number not for citation purposes)
BMC Microbiology 2007,
7:12
response to environmental conditions and using of differ-
venting restriction and ligation reactions. Thus, once
ent resources. The sequenced genome analysis of
T. ther-
inserted into a donor plasmid the selected DNA does not
mophila once more illustrates the complexity of this single
need to be subcloned. It is obvious that this is of high rel-
cell eukaryotic micro.
evance in cloning very large or AT-rich sequences. In thisstudy we present for the first time a
Cre-recombinase
In order to get more insights into functional aspects of the
dependent vector system for ciliates. It allows the inde-
T. thermophila genome molecular biology tools are neces-
pendent construction of expression cassettes on the one
sary that allow the easy handling of the
T. thermophila
and the preparation of acceptor vectors with integration
genes to form the basis of the postgenomic age of this
sites on the other hand. In a second step expression cas-
model organism. The nuclear dimorphism (MIC and
settes can be easily shifted from the donor plasmid into
MAC) of the ciliates offers different possibilities of manip-
various acceptor backbone constructs.
ulating the organism's prop]. However, alteringthe phenotype ultimately needs direct or indirect genetic
Here we describe the proof of concept of such a system for
engineering of the vegetative MAC. The first approaches
the
T. thermophila expression host by two independent
were based on the use of plasmids that take advantage of
examples. We used the C-terminus of a merozoite surface
the vast amplification of the rDNA gene during
anlagen/
antigen (MSP-1) from
P. falciparum and a novel blastici-
MAC development []. However, the episomal presence
din resistance gene (bsdR). They were cloned into the
of these plasmids depends on the presence of antibiotics
donor vector and the gene cassettes were transferred
via
in the culture medium and the plasmids often recombi-
the
Cre-recombinase into different expression vectors.
nate homologously and non-directionally into the endog-
Finally we showed the production of the foreign proteins
in the ciliate
T. thermophila.
The stable integration of expression or knock out cassettes
into the diploid MIC provides a second method to manip-
The goal of this work was to establish a system that facili-
ulate the ciliate's genome, because after conjugation of
tates the cloning and then allows a flexible shuttling of the
two different mating types the old MACs disintegrate and
corresponding sequences and/or expression cassettes into
new ones form that carry the new information derived
the appropriate vector systems. To reach this aim we con-
from the recombinant MIC. The advantage is that one
structed a set of vectors that take advantage of a
Cre-
obtains stable clones that can be crossed
via classical Men-
dependent recombinase system [
delian genetics to combine various properties of different
T. thermophila strains. But this approach is elaborative and
First we constructed the donor plasmid. We selected a
time consuming. Furthermore, it has recently been shown
pCR-TOPO vector as backbone and removed the ampicil-
that scan RNAs (snRNA) derived from the old MAC epige-
lin resistance (ampR) gene by BspHI digestion and subse-
netically control the genome rearrangement of the new
quent religation of the plasmid. In a second step a 1.8 kb
developing MAC [. Thus this RNAi-like mecha-
DNA cassette was inserted by using EcoRI sites. This artifi-
nism may cause problems due to partial deletion of for-
cial cassette (K42) has a modular structure and contains a
eign expression cassettes in the developing new MAC.
histone promoter (
H4-1), a signal peptide (encoding thefirst 39 aa of
P. falciparum surface protein MSP1) fused to
So far ciliate expression vectors rely on large double rDNA
the EYFP reporter protein and a histidine stretch (6xH) as
origin stretches to ensure a stable propagation in
T. ther-
well as the beta tubulin terminator
(BTU2) of
T. ther-
mophila cells or on large flanking integration sites of non-
mophila. All of these DNA modules can be easily changed
coding regions that are necessary for a proper and efficient
by using unique restriction sites. The whole cassette is
homologous recombination into the gene loci of the MIC
flanked by
loxP sites on the 5' and 3' ends (see Additional
or MAC. In both cases the AT-richness of these functional
for basic donor plasmid see figu
DNA sequences cause problems in handling and cloning.
A chloramphenicol resistance (CmR) was inserted
Recombinases like
Cre, Flp or the λ system catalyze rear-
between the
loxP site and the
BTU2 terminator to reduce
rangements of DNA at specific sequences ]. This
background clones, because this CmR is only translated in
enables the insertion of mobile DNA elements into the
E. coli if a correct site-specific recombination between
host genome. Consequently these recombinase mecha-
acceptor and donor plasmid has been occurred. However,
nisms were used to develop different systems that simplify
due to the modular architecture of the pDL-plasmids also
the molecular genetic applications. From the technical
other resistance markers can be used for this purpose (
e.g.
point of view these techniques allow the flexible and fast
tetracycline, zeocin etc.). In a next step, the
sacB marker
transfer of DNA sequences from donor plasmids into
gene was inserted into the intermediate vector. The
sacB
multiple adequate acceptor backbones thereby circum-
gene product metabolises sucrose into levansucrose, a
(page number not for citation purposes)
BMC Microbiology 2007,
7:12
toxic substance for
E. coli cells. The parallel usage of the
(
H4-1-MSP119-
BTU2) was ligated into the pre-cut pH4T2
Cm resistance (selection) and the
sacB (counter-selection)
vector to obtain an rDNA-based MSP119 expression plas-
gene strongly inhibits the presence of non-recombinant
mid. An analogous approach was done with the pKOI
clones. Finally, we replaced the Enhanced Yellow Fluores-
plasmid. Eight clones were randomly picked and analyzed
cence Protein (EYFP) cDNA by the MSP119 cDNA from
P.
by restriction analysis. All of them (8/8) were negative and
falciparum or the blasticidin resistance gene (bsdR) to
most of the pH4T2 backbones were degraded or frag-
demonstrate that the whole system facilitates cloning and
mented during the ligation, transformation selection and
expressing foreign genes like previously shown for other
propagation procedure. The supplementary figure 1 illus-
host systems
e.g. arabidopsis.
trates a typical result of such an approach. In most casesonly 2–3% (one of 30 to 50 clones) carries the complete
Recently, we described the bifunctional dihydrofolate
expression cassettes in a complete plasmid.
reductase and thymidylate synthase (DHFR-TS) of
T. ther-mophila. Both enzyme activities play a crucial role in DNA
Extracts of cells that were transformed with the pKOIX-
synthesis. The loss of these essential activities can be used
MSP119 plasmid were analyzed for expression of a 19 kDa
as an auxotrophic marker in
T. thermophila. We developed
protein fragment of the MSP1 protein. We used cell
a vector system that combines the
knock out of the endog-
extracts of four independent stably transformed strains
enous DHFR-TS gene with the
knock in of an expression
and compared them to the non-transformed 1868/7
cassette that encodes a foreign gene (p. Appro-
wildtype strain. In all tested cell extracts the recombinant
priate acceptor vectors for the
T. thermophila system were
19 kDa fragment of MSP1 (MSP119) could be detected by
created by cloning the
loxP-promoter site (loxprom) into
the specific monoclonal antibodies (mAb2.2, mAb7.5,
this pKOI vector backbone as well as into a previously
mAb12.8 and mAb12.10 were kindly provided by Prof.
described rDNA based episomal plasmid (pH4T2). The
McBride, Edinburgh, UK) the wildtype nega-
new vectors were named into pKOIX (pKOI backbone)
tive control no signal could be found. The results are sum-
and pAX (pH4T2 backbone). A scheme of the acceptor
marized In previous expression experiments
vector structure is given in figure
we could also demonstrate that the rDNA plasmid is capa-ble of expressing the MSP1 C-terminus (data not shown).
The final expression vectors pAX-MSP119/pAX-bsdR andpKOIX-MSP119/pKOIX-bsdR were generated
via the novel
We attempted to confirm the
Cre-recombinase dependent
recombinase mechanism. In general, a mixture of 100 ng
cloning by using a second independent expression mod-
of donor and 100 ng of acceptor plasmid yielded 30 to 50
ule. Therefore pAX and pKOIX constructs that carry the
initial positive clones that were able to grow on LB-agar
bsdR expression cassette (pAX-bsdR, pKOIX-bsdR) were
plates supplemented with both chloramphenicol and
transformed into conjugating and vegetative
T. ther-
mophila wildtype strains according to protocols previouslydescribed. The transformants (clone1- clone10) were cul-
We picked six clones of the MSP119 recombinase reactions
tivated in SPP-medium supplemented with thymidine
and analyzed them by diagnostic PCR and restriction
and increasing concentrations of paromomycin (figure
analysis to test the efficiency of the novel recombinase
o ensure a stable propagation of the clones. The same
approach. All of the analyzed clones were positive (6/6).
clones were cultivated in SPP-medium without antibiotics
This finding was independent of the used acceptor plas-
(figure n a second step we performed a blasticidin
mid. The recombinant pAX- as well as pKOIX-plasmids
growing assay and switched the antibiotic from paromo-
carry complete expression cassettes and an all complete
mycin to blasti) or applied both antibiot-
plasmid backbone. The results are shown in
ics in parallel. In all experiments the wildtype (WT) that
left column illustrates the results using the pAX and right
did not contain a resistance gene died within 2–5 days. As
column the pKOIX backbone. Positive clones were ana-
expected the mock transformant (MT) that only carried
lyzed by restriction analysis (XhoI and SacI) and diagnos-
the
neo2 cassette (resistance against paromomycin) died
tic PCR (360 bp fragment in positive clones), verifying a
when blasticidin was added to the SPP-medium (bsd anti-
correct recombinase event. In general nearly all clones
biotic control).
(80–100%) obtained were positives and have a completebackbone. We obtained such quotes in all performed
Cre-
Interestingly, we observed that the pKOIX-bsdR trans-
recombinase reactions (data not shown).
formants were more stable when compared to the pAXclonesD). Eight out of the ten analyzed inde-
We compared the recombinase efficiency of generating
pendent pKOIX-bsdR clones are resistant against both
recombinant expression plasmids standard cloning tech-
antibiotics. In contrast to that only 30% of the pAX
niques. The pDL-MSP119 (see Addi was
clones (3/9) displayed both resistances. This is probably
digested with NotI and SacI and the corresponding insert
due to recombination events between the rDNA plasmids
(page number not for citation purposes)
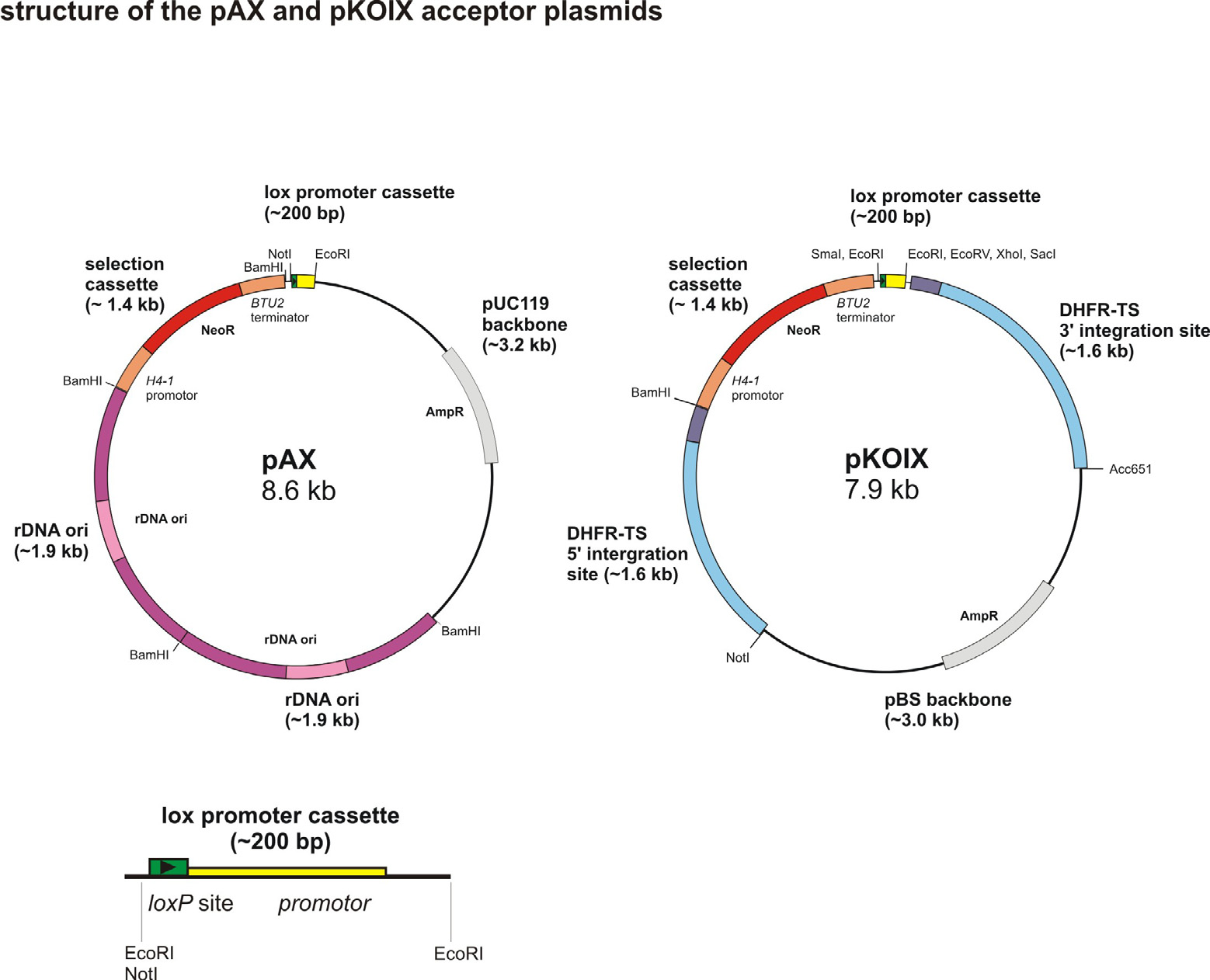
BMC Microbiology 2007, 7:12
The acceptor vect
ors pAX and pKOIX
The acceptor vectors pAX and pKOIX. Structure of pKOIX and pAX acceptor plasmids. The loxprom site that consists
of a loxP site (green) and the bacterial promoter (yellow) was inserted into pH4T2 by using the NotI and EcoRI and into pKOI
by EcoRI restriction sites, respectively. The arrow head in the loxP sites indicates the loxP orientation. The gray/black parts cor-
respond to the backbone sequences that are necessary for propagation in E. coli, and the red part of both acceptor vectors
illustrate the neo2 selection cassette for selection in T. thermophila by paromomycin. The purple colored part of pAX illustrates
the AT-rich double rDNA ori which is necessary for the propagation in T. thermophila. The blue part in pKOIX shows the AT-
rich 5' and 3' integration sequences (bright blue, up/downstream and dark blue coding sequences). They are part of the endog-
enous DHFR-TS gene locus.(Bottom: detail of loxprom site).
backbones and/or due to the very similar architecture of
the neo2 and bsdR resistance cassett). In
Protozoan and functional genomics are an exciting
summary these results illustrate that the Cre-dependent
research area. More and more genomes of eukaryotic
modular donor plasmids in combination with the
microorganisms have already been completely sequenced
recently described knock out/knock in concept (pKOIX)
(e.g. Plasmodium falciparum, Tetrahymena thermophila, Par-
provides an easy and sustainable system to establish a
amecium tetaurelia) or will be available soon (e.g. genomes
resistance testing tool.
from species of Toxoplasma or Entamoeba-is
(page number not for citation purposes)
BMC Microbiology 2007, 7:12
he Cre-recombinase reaction with novel donor and acceptor plamids
Analysis of the Cre-recombinase reaction with novel donor and acceptor plamids. Results of the Cre-recombinase
reactions using pKOIX, pAX, and pDL-MSP119, respectively. Left column: Analysis of recombinant pAX-MSP119 clones. Top: The
restriction analysis of recombinant pAX clones with XhoI and SacI and are shown. The analyzed positive clones (1–6) have an
8.6 kb backbone and a 1.2 kb insert. The vector backbone without the insert (control B) and the donor plasmid insert (MSP1 )
are shown in lanes B and I. Bottom: The diagnostic PCR confirms that all analyzed clones are positive, because the amplification of the specific 360 bp fragment is only possible in recombinant clones. The acceptor control (A) is negative and the fragment in the donor control (D) is an unspecific artefact (200 bp). Right column: The analogous approach as shown in the left column with the acceptor plasmid pKOIX instead of pAX. The analyzed positive clones carry a 7.9 kb backbone and the 1.2 kb insert. The additional band at 1.0 kb is due to a SacI site in the Cm resistance. Lanes B and I show the used backbone (pKOIX) and the 1.2 kb insert (MSP1 ). The diagnostic PCR leads to the specific 360 bp fragment like in the pAX-MSP1 approach. The 200 bp
fragment in the donor control, (D) is an unspecific artefact. The acceptor control (A) is negative.
allows new insights into evolutionary mechanisms as well
lian cell lines, Drosophila, yeast or E. coli these tools have
as the discovery of new biochemical pathways or the iden-
been developed and optimized for decades. In contrast to
tification of promising vaccine candidates against patho-
this some tools are lacking to deal with unusual properties
genic protozoans.
and pitfalls of unusual organisms. AT-rich genomes forexample cause difficulties in handling the DNA
The recently characterized genome of the ciliate Para-
sequences. Also the AT-richness of the T. thermophila
mecium tetaurelia for example elucidated that three succes-
genome causes the main challenge in altering the ciliates
sive whole genome duplications lead to nearly 40,000
phenotype. Previously described episomal expression
genes, illustrating that these mechanism allows a an excel-
plasmid consist of a pUC backbone that enables propaga-
lent adaptation to environmental cond.
tion in E. coli, two 1.9 kb parts forming the rDNA ori andthe neo2 cassette that allows the selection of the trans-
However, functional genomics and subsequent proteomic
formed ciliates. The empty vector is sized about 8.4 kb.
studies require tools that allow the analysis and manipu-
Especially the two rDNA origins are (3.8 kb) AT-rich
lation of certain genes of interest. For the most common
sequenc sequences are probably one reason
model organisms and expression systems like mamma-
why this vector tends to recombinate into the highly
(page number not for citation purposes)
BMC Microbiology 2007, 7:12
expression cassettes. This indicates that the AT-rich DNAof the double rDNA or of the integration sites is responsi-ble for the described problems. There is a demand for sim-plification of the genetic manipulation of AT-richprotozoans. Therefore both available vector systems wereoptimized.
Creating and tuning of expression cassettes in a small andflexible donor vector and the subseqeuent construction offinal expression vectors by an easy and robust shuttling ofthe expression module provided a solution to this prob-lem. The constructed donor vector possesses a modularstructure and is small sized. It lacks AT-rich sequences likerDNA or integrative sites. Thus the gene of interest as wellas signal peptides, promoter and terminator sequencescan easily be substituted via unique restriction sites. Thisoffers the possibility to establish simple test systems likethe here shown resistance gene test system. Furthermore,different constitutive, cell cycle dependent or induciblepromoter sequences can be combined to various genes ofinterest.
The whole DNA cassette of the donor vector is flanked byloxP sites to enable the Cre-dependent site-specific transfer
Expression of the 19
kDa C-terminus of MSP1 in T. ther-
into appropriate acceptor plasmids.
Expression of the 19 kDa C-terminus of MSP1 in T.
thermophila. Aliquots (extract of 50,000 cells) of a non-
transformed wildtype and cells of transformed clones (clones
As acceptor plasmids we used both, the backbone of the
1–4) that were transformed with pKOIX-MSP1 construct
episomal rDNA plasmid pH4T2 and the recently
were separated on 15 % SDS-PAGE. The recombinant 19
described pKOI backbone. Up to now, these concepts and
kDa fragment of MSP1 could be detected in all clones (1–4)
the paclitaxel system developed by Gaertig et al. are the
with four independent monoclonal antibodies (mAbs 2.2, 7.5,
only known expression vector concepts that are available
12.8 and 12.10) all derived against the C-terminus of MSP1 of
for the T. thermophila system [ only
P. falciparum. No signal could be observed in the wildtype
observe a complete transfer but also an expression of the
negative control. The monoclonal antibodies were kindly
encoded genes of the shuttled expression module (bsd
provided by Prof. J. McBride (Edinburgh, UK).
resistance gene and the C-terminus of the MSP1 protein),illustrating the high efficiency of the new system.
amplified endogenous rDNA chromosomes of the host
The ciliates are one of three evolutionary lineages that
make up the alveolates. The two further groups are dino-flagellates and apicomplexans. Especially the endopara-
Recently, we developed a knock out/knock in system
sitic apicomplexans contain a number of human and
(pKOI) that is based on the stable integration into the
animal pathogens (e.g. the genera Plasmodium, Toxo-
endogenous gene locus of the DHFR-TS. The main advan-
plasma, Cryptosporidium (for details see the apicomplexans
tage of this concept is that a stable knock out can be moni-
databasehe most important protozoans from this
tored by the complete loss of the DHFR-TS activity,
apicomplexans group are Plasmodium species that causes
resulting in an auxotrophy for thymidine. Thus this
marker system allows the propagation of recombinant T.
thermophila cells without rDNA ori sequences [How-
Intriguingly, the genome of Plasmodium falciparum is the
ever, large AT-rich stretches are necessary to ensure good
most AT-rich genome known so far (for details see Plas-
integration efficiency by homologous recombination. In
modium database [. This illustrates that the han-
the case of the pKOI constructs a 1.6 kb regions of the 5'-
dling of (large) AT-rich DNA sequences is not a problem
and a 1.6 kb 3'-region of DHFR-TS gene have been added
limited to T. thermophila applications. However, we and
to the vector backbone. The uptake of ligation reactions
others demonstrated that the T. thermophila expression
and the subsequent amplification in E. coli often resulted
system is able to express proteins from the malaria para-
in reduced and fragmented backbones and the loss of the
site Plasmodium faciparum, suggesting that this "distant
(page number not for citation purposes)
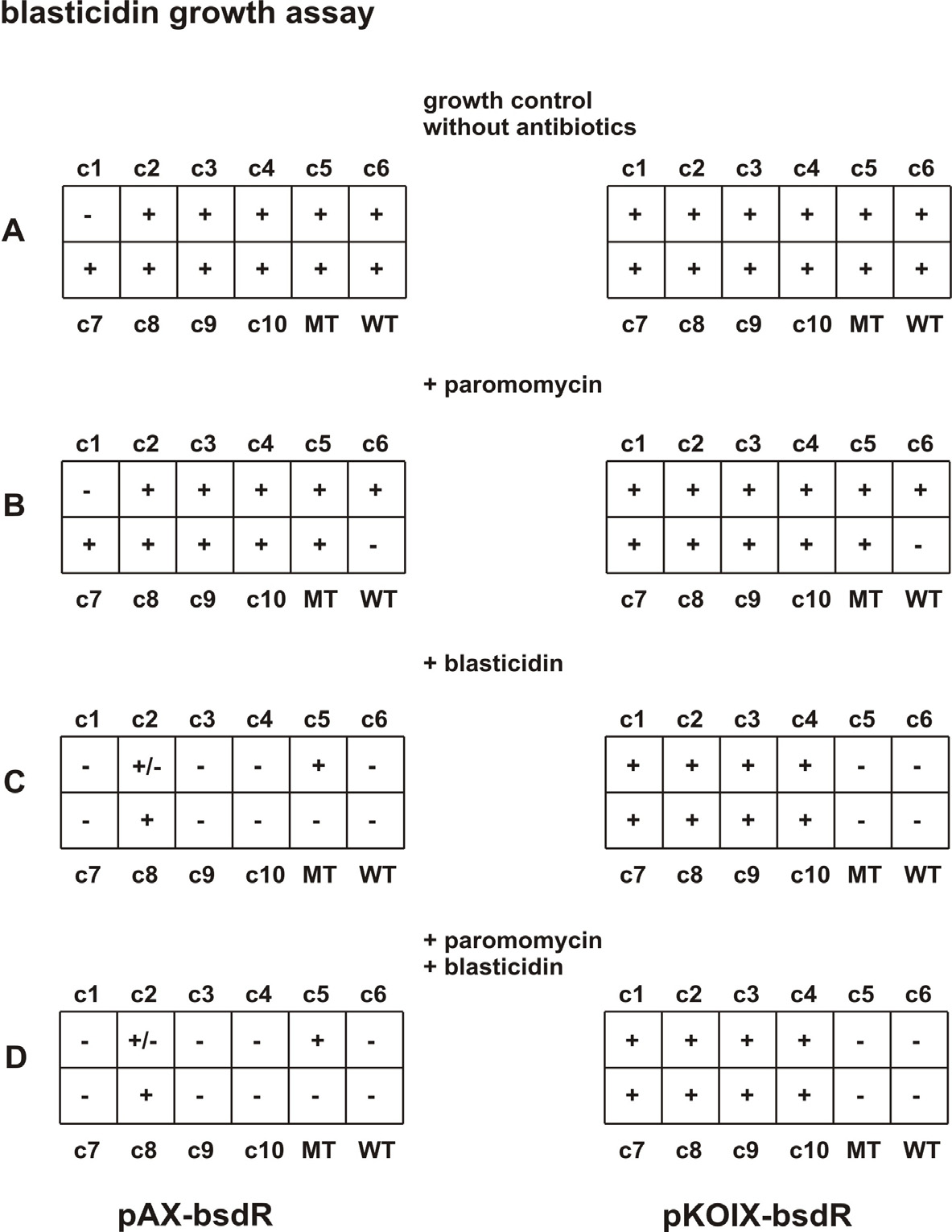
BMC Microbiology 2007, 7:12
Blasticidin growth assay
Figure 4
Blasticidin growth assay. We created a donor plasmid – pDL-bsdR – that encodes the blasticidin resistance gene under the
control of the H4-1 promoter and the BTU2 terminator (see figure 5B). It has a similar structure as the previously described
neo2 cassette (resistance against paromomycin) that is also present in the here used acceptor plasmids pAX and pKOIX. Using
the Cre-dependent recombinase we generated the expression plasmids pAX-bsdR and pKOIX-bsdR and transformed T. ther-
mophila strains. Blasticidin resistance assay: left column: clones of cells transformed with pAX-bsdR, rightcolumn: clones of cells
transformed with pKOIX-bsdR. Several independent clones (c1 to c10) were tested for bsd resistance. As controls we used
the wildtype strain 1868/7 (WT) and a mock transformant (MT) that only carried the neo2 resistance gene A: growth control
of clones in SPP-medium without antibiotics; B: same clones as presented in A selected in SPP-medium with 400 μg/mL paro-
momycin after 5–10 days C: same clones as in B, but cultivated for 3–5 days in SPP-medium with 100 μg/mL blasticidin; D:
identical clones as in B/C cultivated in SPP-medium for 3–5 with both antibiotics, paromomycin (400 μg/mL) and blasticidin
(100 μg/mL). + strong growth of clones. +/- cells alive, less growing. - no growth, cells died within 2–3 days.
(page number not for citation purposes)

BMC Microbiology 2007, 7:12
Figure 5 of the used donor plasmids and expression cassettes
Structure of the used donor plasmids and expression cassettes. A: Scheme of the basis donor plasmid (linear struc-
ture) with its modular structure. The cassette allows the simple substitution of promoter, signal peptide (sp) gene of interest
and terminator DNA sequences without loosing the flanking loxP sites due to unique restriction sites. B: "Floxed" (flanked by
loxP) expression modules were used that encode an expression cassette for the MSP1 C-terminus gene (top) and the blasticidin
resistance (bsdR, bottom) gene, respectively. All expression cassettes are flanked by a histone promoter (H4-1) and a beta
tubulin terminator (BTU2).
cousin" can serve as an expression system for proteomic
ical tool allows a more flexible and easy handling of DNA
applications of other protozoans ]. Furthermore, the
sequences and might reveal a concept that can easily be
here shown vectors can be modified to enable an adapta-
transferred to other protozoan host systems.
tion to optional requirements. For example, the DHFR-TSintegration sites of pKOI can be substituted by DNA
sequences of other species to allow an integration into
The here presented Cre-dependent recombinase system is
other host systems (e.g. Plasmodium or Paramecium spe-
the first one that has been established for a protozoan sys-
cies). Thus the here presented powerful molecular biolog-
tem. It allows a facilitated shuttling of DNA into expres-
(page number not for citation purposes)
BMC Microbiology 2007, 7:12
sion vectors and thereby an easier handling of these AT-
Both cDNAs were restricted (BglII or BamHI) to insert the
rich DNA sequences.
fragments into the EcoRV/BglII pre-cut, linearised pDL-K42 vector. All constructs were verified by sequencing
As the concept can easily be adapted to further unicellular
(Carpegen, Muenster, Germany).
eukaryotes the whole system provides a powerful molecu-lar biology tool in protozoan research.
Acceptor plasmidsEpisomal acceptor plasmid
We amplified the lox-promoter cassette by using the primers
loxprom F/R: (5'-AGTCTgaattcACGTCAGGTGGCACTTTTC-
Donor plasmid
3') and (5'-TTgaattcgcggccgcATAACTTCGTATAG-3'). As tem-
The construction of the donor plasmid was done in sev-
plate we used the lox-promoter site of the acceptor vector
eral steps. Firstly, a pCR-TOPO backbone was restricted
pLP GADT7 (BD Clontech, Heidelberg). The small letters
with BspHI and re-ligated to eliminate the ampicillin-
correspond to the used restriction sites within the
resistance (ampR). Secondly, this smaller sized backbone
sequence. The corresponding loxprom PCR fragment was
that only carries the remaining kanamycin resistance
digested with EcoRI and NotI and cloned into a previously
(kanR) was digested with EcoRI to insert the K42 cassette
described double rDNA ori based pH4T2 shuttle plasmid
(made by Geneart, Germany). This artificial K42 expres-
]. This new acceptor plasmid was named pAX.
sion cassette encodes the first 39 amino acids of the MSP1sequence, fused to enhanced yellow fluorescence protein
Integrative acceptor plasmid
(EYFP) and to a histidine tag. The expression of this gene
The recently described knock out construct pKOI that
cassette is controlled by a histone promoter (H4-1) and a
allows the integration into the dihydrofolate reductase
beta tubulin terminator (BTU2). The whole K42 cassette is
thymidylate synthase (DHFR-TS) gene locus of T. ther-
flanked by two loxP sites ("floxed") of the same orienta-
mophila was used to construct a second acceptor vector
tion. In the third step a chloramphenicol resistance gene
]. We used the unique EcoRI site to insert the lox-pro-
(CmR) was amplified with the primers CmRF: (5'-
moter site. The loxP orientation promotes a site-specific
insertion in the same orientation as the neo2 resistance
and CmRR (5'-TTTgagctcggccggccAAATTACGCCCCGCCCT-
cassette of pAX and pKOIX. Details on primers donor and
GCCACTC-3') and inserted into the K42 sequence by using
acceptor sequences and the constructs are available at Cil-
SpeI and SacI. The CmR gene is localized between the BTU2
ian AG (Muenster, Germany).
terminator and the down stream loxP site. In a fourth stepthe sacB gene from Bacillus subtilis was amplified by using the
Cre-recombinase reaction and selection in E. coli
primers pair sacBF/R (5'-TTactagtACATATACCTGCCGTTCAC-
Donor plasmids pDL-bsdR and pDL-MSP119 and acceptor
TATTATTTAGTG-3') and (5'-TTactagtGGCATTTTCTTTT-
plasmids pKOIX and pAX were amplified in LB-medium
GCGTTTTTATTTGTTAACTGTTAATTGTCC-3'). This sequence
containing kanamycin or ampicillin, respectively. The
was inserted by using a unique SpeI site of the intermediate
recombinase reactions were performed as follows: Aliq-
donor vector site of the third step. The sacB sequence is nec-
uots of 100 ng donor and 100 ng acceptor plasmids were
essary to allow a counter selection by adding sucrose to
mixed in reaction buffer. Cre-recombinase was added and
the LB-agar plates. For amplification of the CmR and sacB
samples were incubated for 25 min at 25°C. The reaction
sequences a pDNR was taken as template (data not
was stopped by heat inactivation for 10 min at 70°C. Aliq-
shown). The combination of the CmR and sacB gene is
uots of the reaction were purified by using the Montage
similar to the previously established Creator (BD Clon-
Kit (Millipore, Schwalbach) and transformed into E. coli
tech, Heidelberg) concept. Finally, the intermediate vector
strain DH10B (Invitrogen, Heidelberg). Positive clones
pDL-K42 was digested with EcoRV and BglII to replace an
were selected on LB-agar plates supplemented with ampi-
EYFP insert by a cDNA encoding the 19 kDa C-terminus
cillin, chloramphenicol and 7% sucrose. Recombinant
of the merozoite surface antigen 1 of Plasmodium falci-
pKOIX and pAX plasmids were propagated in LB-medium
parum (MSP119) and the cDNA of the blasticidin resist-
containing ampicillin. First the clones were analyzed by
ance gene (bsdR), respectively. The bsdR cDNA was
diagnostic PCR by using the primers PCP1 (5'-GCTCAC-
amplified using the primers bsdF/R (5'-gatatcAT-
CGTCTTTCATTGCC-3') that binds in the Chloramphenicol
GGCCAAGCCTTTGTCTCAAG-3') and (5'-ttagatct
resistance gene and PCP2 (5'-TCCGCTCATGA-
TCAGCCCTCCCACACATAACCAGAGG-3'). The MSP119
GACAATACC-3') that binds in the prokaryotic Chloram-
cDNA was amplified from a genomic DNA preparation of
phenicol promoter region. Both elements are only
P. falciparum 3D7 (kindly provided by Prof. Lanzer Hei-
connected by a successful recombinase reaction, leading
delberg, Germany) by using the primers MSP119-F (5'-
to a specific PCR product (ca. 360 bp). Clones were ana-
AACATTTCACAACACCAATGCG-3') and MSP119-R (5'-
lyzed using the restriction enzymes XhoI and SacI to verify
whether or not the backbone was complete.
(page number not for citation purposes)
BMC Microbiology 2007, 7:12
Ciliate strains, cultivation and transformation
room temperature. After washing with PBS/T for 30 min
Tetrahymena thermophila strains B 1868/4, B 1868/7 and B
we applied the second antibody (HRP-conjugated goat
2068/1 were cultivated in skimmed milk medium (2%
anti mouse serum; dilution 1:1000) in PBS-TM. The blots
skimmed milk, 0.5% yeast extract, 0.1% ferrous sulphate
were developed by chemiluminescence using SuperSignal
chelate solution and 1% glucose) in SPP-medium (0.5%
West Femto Max Sensitivity Substrate (Pierce Biotechnol-
proteose peptone, 0.5% yeast extract, 0.1% ferrous sul-
ogy) in combination with conventional X-ray film devel-
phate chelate solution and 1% glucose) or in modified
CDM (modified from Hellenbdium asdescribed previously [. We used vegetative growing
non-conjugating T. thermophila strains. The transforma-
Most experiments, the concept and the manuscript were
tion of the T. thermophila cells was performed as described
made by TW, AR generated the intermediate plasmids and
previously. Transformed cells were distributed on 96 well
UB transformed the ciliates and selected the positive
plates. Positive individual clones were isolated (single cell
clones. LH participated in construction of the pKOI back-
isolation) and further cultivated in 24 well plates.
bone and MWWH in the conceptual work. Vectors andstrains can be made available upon request from MWWH.
Selection, allelic assortment and DHFR-TS knock out assay
All authors read and approved the final manuscript.
T. thermophila cell proliferation assay: For the first 16 hafter biolistic bombardment transformants were grown in
skimmed milk medium. After that transformed cells weregrown on SPP-medium with increasing concentrations of
Additional File 1
paromomycin (from 100 μg/mL to 1000 μg/mL) to sup-
Construction of the first donor plasmid. A: Scheme of the used modular
port the allelic assortment process. After 2–4 weeks each
artificial cassette K42. It allows the substitution of promoter, gene of inter-
clone was cultivated on CDM replica plates with or with-
est and terminator sequences without losing the flanking loxP sites. B:
out thymidine (10 mg/mL). Functional DHFR-TS knock
The "floxed" artificial sequence was inserted into a pCRTOPO backbone,
out clones are only able to grow in CDM medium supple-
lacking the ampicillin resistance gene. In the next steps a chloramphenicol
mented with thymidine. The viability of the DHFR-TS
resistance and a sacB counter-selection cassette were added. This basic
knock out strains was monitored by determining the
donor plasmid was used to replace the EYFP cDNA (see figureClick here for file
growth kinetic as previously described in more .
Blasticidin growing assay
Clones that were transformed with pAX-bsdR or pKOIX-
Additional File 2
bsdR plasmids were first selected in SPP-medium supple-
Recombination and fragmentation of large AT-rich plasmids. This figure
mented with paromomycin and thymidine. After that the
illustrates the undesired recombination events that lead to fragmented
presence of the bsd resistance cassette was verified by add-
plasmids during the standard cloning procedure (ligation, transformation, selection and propagation) in E. coli. M: marker; 1 kb ladder (generuler,
ing 100 μg/mL blasticidin to the CDM/SPP-media. Posi-
MBI Fermentas, 1–8: Analyzed clones; B/I: backbone DNA (8.4 kb) and
tive clones are resistant against the bsd addition to the
insert DNA (ca. 1.2 kb) that was used for the ligation reaction.
culture medium, negative clones died within two to three
Click here for file
days. Both pAX-bsdR as well as pKOIX-bsdR clones are
able to grow in SPP/CDM-media with both antibiotics in
the medium (400 μg/mL paromomycin and 100 μg/mLblasticidin).
SDS-PAGE and Western blot
SDS-PAGE and Western blot analysis were done as previ-
ously described ,]. Briefly, aliquots (50,000 cells) of
transformed cells were resuspended in sample buffer
boiled for 10 min and separated on 15% SDS-PAGE. The
gels were blotted onto nitrocellulose membranes and
blocked of 1 h at room temperature (or at 4°C overnight)
in PBS containing 0.05% Tween 20 and 5% skimmed
milk (PBS-TM). The expression of recombinant MSP119 in
transformed ciliates was detected by specific monoclonal
anti MSP119 antibodies kindly povided by Prof. McBride
(Edinburgh, UK). The monoclonal antibodies were
diluted 1:100 in PBS-TM and incubated for 1 hour at
(page number not for citation purposes)
BMC Microbiology 2007, 7:12
Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den
We would like to thank Prof. Jana McBride for the monoclonal antibodies
against the C-terminus of the MSP1 protein and Prof. Michael Lanzer for
Methods Enzymol 2000, 328:575-592.
providing us P. falciparum genomic 3D7 DNA. Furthermore we would like
to thank Dr. Ingo Aldag for critical reading the manuscript and Linsay Hue-
Annu Rev Biochem 1989,
bers for excellent technical assistance. This research was supported by the
Herrmann L, Bockau U, Tiedtke A, Hartmann MW, Weide T:
TIP grant No. 005-0303-0003 from the Ministry of Economics Affairs and
Energy of the state of North Rhine Westfalia (NRW, Germany) to Cilian
BMC Biotechnol 2006, 6:21.
Wilson CF, Anand R, Clark JT,
Collins K, Gorovsky MA: Curr Biol
Parasite Immunol 1987,
Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR,
McBride JS, Heidrich HG:
Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salz-
berg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith
RK Jr., Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC,
Biochem Parasitol 1987, 23:71-84.
Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart
Protozoan Genomes 2007
BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaer-
tig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron
Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B,
NJ, Cherry JM, Stover NA, Krieger CJ, del TC, Ryder HF, Williamson
Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc
SC, Barbeau RA, Hamilton EP, Orias
I, Bouhouche K, Camara F, Duharcourt S, Guigo R, Gogendeau D,
Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le MA, Lepere G,
PLoS Biol 2006, 4:e286.
Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Ser-
Turkewitz AP, Orias E, Kapler G:
rano V, Zagulski M, Dessen P, Betermier M, Weissenbach J, Scarpelli
Trends Genet 2002,
C, Schachter V, Sperling L, Meyer E, Cohen J, Wincker
Cech TR: Biochem Soc Trans 2002,
Nature 2006, 444:171-178.
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carl-
Asai DJ, Wilkes DEJ Eukaryot
ton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA,
Microbiol 2004, 51:23-29.
Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shal-
Asai DJ, DeWall KM, Lincoln LM, Smith RK:
lom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft
Methods Mol Biol 2001, 161:269-278.
D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ,
Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY,
Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM,
Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis
RW, Fraser CM, Barrell B:
Cell 1996, 84:843-851.
Nature 2002,
Yao MC, Fuller P, Xi X:
Science 2003,
Gaertig J, Thatcher TH, Gu L, Gorovsky MA
Yao MC, Chao JL:
Proc Natl
Acad Sci U S A 1994, 91:4549-4553.
Annu Rev Genet 2005, 39:537-559.
The Apicomplexan Database Resources - ApiDB 2
Wheatley DN, Rasmussen L, Tiedtke A:
The Plasmodium Genome Resource - PlasmoD 2007 [
Bioessays 1994, 16:367-372.
Kiy T, Tiedtke A:
Peterson DS, Gao Y, Asokan K, Ga
Appl Microbiol Biotechnol 1992, 38:141-146.
Gaertig J, Gao Y, Tishgarten T, Clark TG, Dickerson HW:
Mol Biochem Parasitol 2002, 122:119-126.
Hellenbroich D, Valley U, Ryll T, Wagner R, Tekkanat N, Kessler W,
Nat Biotechnol 1999, 17:462-465.
Appl Microbiol Biotechnol
Methods Cell Biol 2000, 62:127-186.
Tetrahymena Genome Database - TGB
Weide T, Herrmann L, Bockau U, Niebur N, Aldag I, Laroy W, Con-
treras R, Tiedtke A, Hartmann
Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J,
BMC Biotech-
Gorovsky MA, Bruns PJ:
nol 2006, 6:19.
Genetics 1997, 146:135-147.
Pan WJ, Blackburn EH Nucleic Acids Res
1995, 23:1561-1569.
Mochizuki K, Gorovsky MA: Curr Opin Genet Dev 2004, 14:181-187.
Abremski K, Hoess R: J Biol Chem 1984, 259:1509-1514.
Sadowski PD: BMC Biotechnol 2003,
3:9.
(page number not for citation purposes)
Source: http://www.cilian.de/download/1471-2180-7-12.pdf
NORTHERN TERRITORY INSTITUTE OF SPORT Northern Territory Institute of Sport Sports Supplement Guidelines (as at 24 May 2013) Overview - This NTIS Sports Supplement Guidelines document provides advice and direction on use of supplements for NTIS athletes, coaches and service providers. The Guidelines follow recommendations made by the AIS Sports Supplement Panel, whose membership includes people
3.02 Understand the functions and disorders of the nervous 3.02 Understand the functions and disorders of the nervous system 3.02 Essential Questions What are the functions of the nervous What are some disorders of the nervous How are nervous system disorders treated? How does the nervous system relate to the body's communication systems?



