Kamagra gibt es auch als Kautabletten, die sich schneller auflösen als normale Pillen. Manche Patienten empfinden das als angenehmer. Wer sich informieren will, findet Hinweise unter kamagra kautabletten.
A novel approach to extract colon lumen from ct images for virtual colonoscopy - medical imaging, ieee transactions on
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 19, NO. 12, DECEMBER 2000
A Novel Approach to Extract Colon Lumen from CT
Images for Virtual Colonoscopy
Dongqing Chen*
, Member, IEEE, Zhengrong Liang
, Member, IEEE, Mark R. Wax, Lihong Li
, Student Member, IEEE,
Bin Li, and Arie E. Kaufman
, Member, IEEE
Abstract—An automatic method has been developed for
ease of performance, degree of patient compliance, expense,
segmentation of abdominal computed tomography (CT) images
and diagnostic accuracy. Among these methods, only optical
for virtual colonoscopy obtained after a bowel preparation of a
colonoscopy and barium enema might be able to examine
low-residue diet with ingested contrast solutions to enhance the
the entire colon. Optical colonoscopy requires intravenous
image intensities of residual colonic materials. Removal of the
enhanced materials was performed electronically by a computer
sedation, takes approximately one hour to perform, has dif-
algorithm. The method is a multistage approach that employs
ficulty in examing the cecum, the most distal portion, and
a modified self-adaptive on-line vector quantization technique
is expensive. Barium enema requires a great deal of patient
for a low-level image classification and utilizes a region-growing
physical cooperation to obtain X-rays at different views, and
strategy for a high-level feature extraction. The low-level clas-
has a low sensitivity. In recent years, virtual colonoscopy
sification labels each voxel based on statistical analysis of its
three-dimensional intensity vectors consisting of nearby voxels.
technology has been developed as an alternative method of
The high-level processing extracts the labeled stool, fluid and air
massive population screening for examining the entire colon
voxels within the colon, and eliminates bone and lung voxels which
for early cancer detection This
have similar image intensities as the enhanced materials and air,
technology uses a computer system to navigate through the
but are physically separated from the colon. This method was
colon model reconstructed from the patient's abdominal CT
evaluated by volunteer studies based on both objective and sub-
jective criteria. The validation demonstrated that the method has
images. It has been shown that this technology is effective in
a high reproducibility and repeatability and a small error due to
imaging colonic polyps as small as 3 mm in diameter
partial volume effect. As a result of this electronic colon cleansing,
All techniques that examine the colon require a clean lumen,
routine physical bowel cleansing prior to virtual colonoscopy may
eliminating residual materials that can falsely be interpreted as
not be necessary.
colonic masses. Prior to any of these examinations, patients
Index Terms—Bowel preparation, electronic colon cleansing,
undergo a bowel cleansing preparation which includes either
image segmentation, virtual colonoscopy.
washing the colon with a large amount of liquids or adminis-tering medications and enemas to induce bowel movements
This bowel preparation is often more unpleasant than theexamination itself. An alternative method of cleansing the colon
COLORECTAL carcinoma is the second leading cause of would be very attractive. In virtual colonoscopy, contrast solu-
cancer-related deaths in the United States with 56 000
tions can be ingested to enhance the image intensities of the
deaths reported in 1998 and an estimated 131 600 new cases
stool and fluid. By applying image segmentation algorithms,
were diagnosed Colonic polyps that are 10 mm or larger
these colonic materials can be virtually removed from the im-
in diameter are considered to be clinically significant, since
ages without the patient undergoing physical bowel washing.
they have a high probability of being malignant Detection
This paper will focus on the development of an automatic seg-
and removal of smaller sized polyps can eliminate over 90% of
mentation method to remove the contrasted colonic materials
colon cancer cases.
for virtual colonoscopy.
Currently available colorectal cancer screening procedures
include digital rectal examination, fecal occult blood test, flex-ible sigmoidoscopy, barium enema, and optical colonoscopy.
II. METHODS AND MATERIALS
These diagnostic tests differ greatly with respect to safety,
A. Bowel Preparation and Imaging Protocol
Manuscript received May 4, 1999; revised August 18, 2000. This work was
Five volunteers were recruited for this study with written con-
supported by the National Institutes of Health (NIH) under Grant CA79180 of
sents. Some of these subjects were used as training samples for
the National Cancer Institute; Grant HL54166 of the National Heart, Lung and
validating the hypothesis of the method and all of them were
Blood Institute; Grant NS33853 of the National Institute of Neurological Dis-order and Stroke; and Established Investigator Award of the American Heart
used for evaluating the performance of the method. The training
Association. The Associate EWditor responsible for coordinating the review of
samples were chosen for an equal distribution in gender with a
this paper and recommending its publication was W. Higgins.
Asterisk indicates
wide age range, see Table I, where three patients were added to
*D. Chen is with the Departments of Radiology, State University of New
include variation in a large population. All volunteers had the
York, Stony Brook, NY 11794 USA (e-mail:
[email protected].).
following bowel preparation. During the day prior to CT scan,
Z. Liang, M. R. Max, L. Li, B. Li, and A. E. Kaufman are with the Depart-
the volunteers took a high fluid, low residue diet for the meals. In
ments of Radiology and Computer Science, State University of New York, StonyBrook, NY 11794 USA.
order to enhance residual colonic materials, they ingested con-
Publisher Item Identifier S 0278-0062(00)10613-5.
trast solutions of 250 cc barium sulfate suspension (2.1% w/v,
0278–0062/00$10.00 2000 IEEE
CHEN
et al.: A NOVEL APPROACH TO EXTRACT COLON LUMEN FROM CT IMAGES FOR VIRTUAL COLONOSCOPY
SUBJECT INFORMATION, WHERE F AND M STAND FOR THE FEMALE AND
MALE, AND V AND P FOR THE VOLUNTEER AND PATIENT
E-Z-EM, Inc.) with each meal and 120 ml of MD-Gastroview
Depiction of the local volume for a voxel.
(diatriuzoate meglumine and diatriozoate sodium solutions) inequal 60 ml amounts during the evening and in the morningbefore the CT scan. All patients used the same bowel prepara-
component analysis (PCA) was then applied to the local
tion with addition of magnesium citrate laxative and bisacodyl
vector series to determine the dimension of the feature vectors
tablets and suppository for physical colon cleansing.
and the associated orthogonal transformation matrix [i.e., the
Prior to acquiring CT images, 1.0 mg of Glucagon was given
Karhunen–Loeve (K–L) transformation matrix]. The PCA on
intravenously in order to reduce colonic motion and spasm, fol-
the datasets of the training samples showed that a reasonable
lowed by introducing approximately 1000 cc of CO through a
dimension of the feature vectors was 5, where the summation
small bore rectal tube to inflate the colon. All CT images were
of the first five principal components' variances was more than
acquired in less than 40 s during a single breath hold. Using a
92% of the total variance.
GE/CTI spiral CT scanner, 5mm collimation with pitch between
It is computationally costly to determine K–L matrix for each
1.5–2.0 : 1, depending upon the span of the colon as determined
dataset. A general K–L matrix was then determined by the training
from a digital scout radiograph, was performed. Scan parame-
samples and used for segmenting all datasets acquired from the
ters included 120 kVp, 180–280 mA (lower mA for volunteers)
same source, i.e., from the same scanner with the same imaging
and field of view (FOV) between 34–40 cm based on the ab-
protocol described previously. To provide evidence for using the
dominal size. The acquired data were reconstructed at 1-mm in-
general K–L matrix for all datasets acquired from the same source,
tervals with a 512
512 array, resulting in 300–450 slices for
the Kolomogorve–Smirnov test was performed which aims
each dataset. Both supine and prone positions were scanned for
to prove that all datasets from the same source could be regarded
validation purpose. For the same purpose, each volunteer was
as a sample set from an identical probability distribution. A supine
also scanned in the following day after the first CT scan and a
dataset was chosen from each of the training samples. The as-
day of low-residue diet, resulting in four datasets.
sociated cumulative distribution function (CDF) was obtainedand denoted by
(some volunteers have
B. Feature Analysis of Image Data
two supine scans acquired in two consecutive days). By utilizingall datasets (both supine and prone) in the training samples, a
To minimize computing time, the voxels outside the body
general CDF, denoted by
, was also computed. This
contour were first eliminated. The remaining is called body
was regarded as the estimation of the source CDF. Then, the
voxels. This was achieved by a boundary-search algorithm
are identical distributions may be
Similar to Markov random field (MRF) models
tested against hypothesis H1:
are not identical,
we assume that a three–dimensional (3-D) object of a
. The differences of greatest magnitude
similar tissue type in a CT image should be in a contiguous
were listed in Table II.
3-D volume, naturally including partial volume effect. It is
The table Table VIII] gives the critical value with a
reasonable to classify the body voxels based on the intensity
two-tail test at a nominal 1% significance level of 0.45. All
similarity within certain spatial range. The diameter of the local
differences listed in Table II are less than 0.45 (the largest
range for a given voxel should be less than 5 mm considering
sample size Table VIII] is 20, where the critical value
the partial volume effect and the 5-mm-thick collimation. By
is associated to this sample size). Hence, we accept H0, i.e.,
the acquisition protocol described above, each voxel was 1
all datasets can be regarded as coming from an identical
mm thick with size in the
– axial plane varying from 0.64
probability distribution. Therefore, the general K–L matrix
to 0.94 mm depending on the FOV. The chosen local volume
determined by the training samples can be applied to segment
is depicted in Fig. 1. Its diameter is less than 4.6 mm in all
all the datasets acquired using the same scanning protocol.
directions. The intensities of those 23 voxels in a local volumeform a twenty–three dimensional (23-D) local intensity vector.
The goal of the low-level processing is to classify the body
C. Vector Quantization Algorithm
voxels based on their local intensity vectors.
For the low-level classification, the K–L transformation was
Each dataset consists of millions of body voxels, where each
first applied to the local vector series. In the K–L domain, the
voxel has a 23-D local intensity vector. This requires inten-
feature vectors were formed by the first five principal com-
sive computational effort to manipulate such a large quantity
ponents from the transformed vector series. Then, the feature
of vectors. To reduce the computing burden, a feature anal-
vectors were classified into several classes. There are several
ysis of the local vector series is necessary The principal
approaches to classify the vectors In general, an automated

IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 19, NO. 12, DECEMBER 2000
THE GREATEST MAGNITUDES OF THE DIFFERENCE BETWEEN THE SUBJECTS= CDF AND THE SOURCE CDF
algorithm is desired, i.e., an unsupervised self-adaptive vectorquantization (VQ) algorithm is a candidate. A self-adaptiveon-line VQ algorithm was developed and is presented below.
Let
, be the feature vector series, where
is the number of feature vectors;
denote the maximum
number of classes; and
be a threshold for vector similarity.
The VQ algorithm generates a representative vector
be the number of feature vectors in the th class
An example showing the intensity value change gradually from tissue
A to tissue B. The overlap area represents the partial volume region.
The algorithm is outlined as follows.
The algorithm is similar to an unsupervised clustering algo-
rithm. The number of classes and the representative vectors are
updated continuously when more vectors are included in the
2) Obtain the class number
calculation. From this point, the algorithm can be regarded as
a learning procedure. It depends only on two parameters:
the upper bound of possible classes and
larity threshold. In abdominal CT images, there are roughly four
classes that can be perceived based on their intensity features:1) air, 2) soft tissue, 3) muscle, and 4) bone or the enhanced
residual materials. The intensity values of these four classes in-
crease from the lowest to the highest. Due to partial volume ef-
Update the representative vector of
fect, there exists an intensity slope between two spatially con-
tiguous tissue areas (see Fig. 2). The voxels in this slope-area
are called partial volume voxels. To mitigate the under or overestimation of tissue boundary, it is reasonable to find the tissue
Generate a new class
boundary within the slope-area rather than on the edge of the
slope-area. In other words, the partial volume area should be di-vided into two subareas. The left partial volume class is calledpartial volume from
and the right one is called partial
3) Label each feature vector to a class
. The partial volume area from
according to the nearest neighbor rule
labeled as tissue
and the partial volume area from
labeled as tissue
in this study. In the CT images, there are two
kinds of voxels within the colon lumen: 1) air and 2) enhanced
materials. Each kind has a partial volume overlap to area of softtissue/muscle. Assigning two partial volume classes to each of
the overlap areas results in four partial volume classes in total.
Therefore, the maximum class number
for the classification
algorithm was set to eight in this study.
is more crucial to the
classification than
. If it is too large, only one class could be
is the Euclidean distance between
obtained. If it is too small, redundant classes may occur. Ac-
gives the integer
which realizes the min-
cording to our numerical experiments,
was set to the square
root of the maximum component variance of the feature vector
The class number and the representative vector for each class
series. This allows the VQ algorithm to achieve the minimum
can be obtained in a single scan on all feature vectors. This re-
class number with the maximum variance. Since T is estimated
duces greatly the computing time as compared to iterative VQ
from the data, the algorithm is self-adaptive.
algorithms, e.g., the LBG algorithm The representativevector of each class is an estimation of the mean vector of that
D. Extraction of the Colon Lumen
class. From the central limit theorem the larger the number
The results of the low-level classification were represented
in a class is, the more accurate the representative vector esti-
as a labeled image with integer values. The colon lumen con-
mates the mean vector of that class. For a colon dataset, there
sisted of four kinds of labeled voxels: 1) air, 2) partial volume
are millions of body voxels. Hence, the representative vector is
from air to soft tissue/muscle, 3) enhanced materials, and 4)
a good estimation to the mean of that class
partial volume from enhanced materials to soft tissue/muscle.

CHEN et al.: A NOVEL APPROACH TO EXTRACT COLON LUMEN FROM CT IMAGES FOR VIRTUAL COLONOSCOPY
ray from an air voxel and examining whether the ray reaches anenhanced voxel within 2 mm.
E. Evaluation Method
Optical colonoscopy is currently utilized as the gold stan-
dard for validating virtual colonoscopy methods for detectingcolonic polyps of 5 mm or larger in diameter. For the electroniccleansing without physical bowel washing, the gold standard isnot applicable. Instead, we acquired both supine and prone scansfor each subject and furthermore repeated the scans on the nextday for the volunteers, aiming to measure the reproducibility,repeatability and robustness on partial volume effect of the pre-sented electronic cleansing technique. The results were subjec-tively judged by an experienced radiologist. Four objective pa-rameters were calculated from the results to indirectly measurethe performance. The first two were used to measure the seg-mentation error created by partial volume effect. The other twowere used to demonstrate reproducibility (by the same dataset)
A CT slice image and the delineated contour of colon lumen.
and repeatability (by both supine and prone scans of the samesubject) of the method. All these four parameters were calcu-lated from the extracted colon lumen.
1) Average thickness of the partial volume layer (ATPV):
These four classes were denoted by 1, 2, 3, and 4, respectively.
This is the average thickness in 3-D space of the partial
By applying the inverse K–L transformation to the class repre-
volume layer from both air and enhanced materials to soft
sentative vectors, the intensities in the original image space for
tissue/muscle. If this parameter is larger than 5 mm, then
each class were obtained. Classes 1 and 3 were easily segmented
polyps of 5 mm in diameter may not be accurately de-
since their intensities were the lowest and the highest, respec-
tively. Since class 1 includes voxels of lung and class 3 includes
2) Partial volume percentage (PVP): PVP
voxels of bone and these voxels are not physically associated
of the volume of partial volume layer from air to soft
with the colon lumen, we can use region-growing strategies to
tissue/muscle and the volume of partial volume layer from
remove the non colon-lumen voxels from classes 1 and 3. This
enhanced materials to soft tissue/muscle)/(the volume of
is the high-level processing.
entire extracted colon lumen).
We removed lung voxels first. The regions of lungs are two
The volume was counted by the number of voxels in the
contiguous 3-D volume on the left and right sides of the chest. If
region. This parameter estimates the ratio of the partial
FOV covers the entire colon, air voxels in the top slice must be
volume voxels to the entire colon lumen.
lung voxels. Two air voxels for both left and right lungs were
3) Mean intensities of the air lumen (AL) and the enhanced
determined as seeds for growing out the entire lung volume
materials (ERM) voxels. The intensity is in HU.
by using a region-growing algorithm. After removing the lung
volume, the remained voxels in class 1 were inside the colon
of the parameter defined previously for the supine dataset
lumen. Given the air lumen, the partial volume voxels of class
is one corresponding to the prone dataset acquired
2 from air to soft tissue/muscle were then determined because
from the same subject on the same day. The definition
they were contiguous to the air lumen in both geometry space
applies to the two-day scans. A smaller value
and intensity feature.
reflects a better reproducibility or repeatability.
Assuming that bone is separated from colon lumen, the voxels
of enhanced materials were then determined from class 3 by a
III. RESULTS AND DISCUSSION
seed in the lumen with the region-growing algorithm. If a voxelbelonged to class 3 and there existed at least one voxel of air
For subjects 3, 4, and 6, only supine scan was acquired (Table
in the lumen which was 2.5 mm or less away from the given
I). For subjects 1, 2, 5, and 7, both supine and prone scans were
voxel, it was a seed for growing out the volume of the enhanced
acquired on two consecutive days. Subject 8 was scanned in the
materials. The partial volume voxels of class 4 from enhanced
supine position on two consecutive days. There was a total of
materials to the soft tissue/muscle were determined by finding
21 datasets, 13 in supine position and eight in prone position.
the class closest to the material class in both geometry space and
The 15 datasets in the training samples were used to determine
intensity feature. Given the four classes of voxels representing
the size of the feature vectors and the general K–L matrix. It
the colon lumen, the last task was to label the boundary voxels
took nearly 6 hours to generate the K–L matrix. This matrix
between air and enhanced materials in the image space. The en-
was then applied to segment all the 21 datasets. Fig. 3 shows a
hanced material forms a basin-like volume with a flat surface
CT slice image and the extracted lumen contour within the CT
due to the gravitation This surface was the boundary be-
image. Fig. 4 depicts the removed enhanced materials in 3-D.
tween air and enhanced material, and was found by sending a
Fig. 5(a) displays the outside view of the entire extracted colon
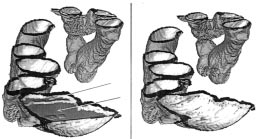

IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 19, NO. 12, DECEMBER 2000
Demonstration of removal of enhanced material (arrows).
The overview of the entire colon lumen (a) and the inside view of a segment of colon lumen (b).
lumen and Fig. 5(b) shows the inside view of a colon segment.
remained on the wall surface near the boundary between
These figures were generated from subject 8. The segmentation
air and enhanced material, these were smaller than 5 mm
of the colon lumen took less than 9 minutes on a SGI/Octane
and were not clinically significant. In one subject, at some
desktop workstation with dual CPU's of R10000, 250 MHz, and
locations, the small bowel adjacent to the colon, appeared
890 MB RAM. No parallel implementation was utilized. Since
"attached" to the colon lumen. For example, subject 1 did
we did not utilize seeded region-growing technique to grow out
not follow the diet instruction, eating a big breakfast on the
the lumen, our method was able to delineate all segments of the
first day prior to CT scan, which resulted in the stomach and
entire colon lumen when collapses presented.
small bowel filled with enhanced materials. The extractedlumen included part of the stomach and several small bowelsegments. (In the second day after correcting the error, thecolon lumen was successfully extracted). For subject 7,
A. Subjective Evaluation
due to a 5-mm thickness collimation, some boundary areas
The radiologist examined the segmentation results and
between the lungs and colon were not resolved, resulted in
was satisfied. The entire lumens from 13 datasets of subjects
connection of lungs and colon lumen. The algorithm could
2, 3, 4, 5, 6, and 8 were successfully delineated, where four
still remove the enhanced materials and extracted the lumen
datasets showed colon collapses. The enhanced materials
including part of the lungs. (A smaller slice collimation
were removed satisfactorily. Although some small artifacts
in data acquisition is recommended). Excluding the two
CHEN et al.: A NOVEL APPROACH TO EXTRACT COLON LUMEN FROM CT IMAGES FOR VIRTUAL COLONOSCOPY
PARTIAL VOLUME ERROR ESTIMATION
REPEATABILITY TEST
datasets of subject 1 and four datasets of subject 7 from the
resolution. This may be achieved by a multidetector ring CT
total 21 datasets, 15 lumens were successfully delineated
and were inspected using our developed virtual colonoscopy
In our algorithm, the colon lumen is delineated by removing
system The attached areas between colon and small
the volume which is not associated with the colon lumen, rather
bowel, as well as between colon and lungs, were deleted
than finding some seeds of the colon lumen to grow out the
manually before navigating through the colon lumens. The
entire lumen. This ensures that the entire colon lumen could be
navigations through both supine and prone scans, as well as
delineated even if there are collapsed segments. This is a very
the second day scans of each subject agreed each other, i.e.,
attractive advantage considering that the colon collapse happens
they agreed with the assumed gold standard that the young
volunteers have no polyps.
The partial volume effect was considered in our algorithm.
The average partial volume layer shown in Table III is 2.57 mm
B. Objective Evaluation
or less in thickness. This ensures that polyps of 5 mm or larger indiameter cannot be affected by the partial volume effect within
The supine and prone datasets acquired on the same day from
the extracted colon lumen. However, some of the flat polyps
subjects 1, 2, and 8, respectively, were used to compute the pa-
could be missed.
rameters for repeatability test and partial volume effect mea-
All the parameter differences in Table IV are less than 8.3%
surement. The results are listed in Tables III and IV, where S#
and most of them are less than 6.5%, demonstrating good re-
means subject #. The reproducibility test from the same dataset
peatability of our method. This statement concurs with the sub-
was excellent, because of the fully automated process.
jective evaluation.
C. Discussion
D. Conclusion
If the bowel preparation instruction was followed, as most
Our two-stage segmentation method designed for the bowel
subjects did, the segmentation results were satisfactory. The en-
preparation using a low-residue diet with ingested colonic
hanced colonic materials were removed successfully except for
contrast solutions was computational efficiency and showed
some small artifacts near the area where air, colon wall, and en-
satisfactory performance. Since the training samples include
hanced material connected. These artifacts form a small artifi-
patient datasets, the segmentation method is applicable to both
cial horizontal ring on the colon wall surface that can be easily
cases with and without additional physical bowel cleansing.
distinguished from the colonic folds. Nevertheless, if a polyp
Most importantly, the electronic colon cleansing technique
smaller than 5 mm is located on the ring, it could potentially
demonstrated the feasibility of performing virtual colonoscopy
be missed in the virtual colonoscopy examination. Further re-
without the need for pre-procedure physical bowel cleansing.
search is needed to minimize these artifacts for detecting smallerpolyps. If a segment of small bowel touches the colon and is
filled with the enhanced materials, this segment may be delin-
The authors greatly appreciate the valuable comments on K–S
eated with a higher probability as the colon lumen. This can be
test from Dr. S. Li.
avoided by not eating any food/drink in the morning prior to CTscan. Another possible solution is to use an interactive display
tool to manually correct the touched segment. The attachment
[1] C. Chatfield and A. J. Collins, Introduction to Multivariate Anal-
of the lungs to colon lumen can be avoided with a higher axial
London, U.K.: Chapman & Hall, 1980.
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 19, NO. 12, DECEMBER 2000
[2] W. Feller, An Introduction to Probability Theory and its Applications,
[12] T. N. Papps, "An adaptive clustering algorithm for image segmentation,"
New York: Wiley, 1968.
IEEE Trans. Signal Processing, vol. 40, pp. 901–914, 1992.
[3] K. Fukunaga, Introduction to Statistical Pattern Recognition, 2nd
[13] T. Parkins, "Computer lets doctors fly through the virtual colon," JNCI,
New York: Academic, 1990.
vol. 86, pp. 1046–1047, 1994.
[4] A. Gersho and R. M. Gray, Vector Quantization and Signal Compres-
[14] G. Rubin, C. Beaulieu, V. Argiro, H. Ringl, A. Norbash, J. Feller, M.
Boston, MA: Kluwer, 1992.
Dake, R. Jeffrey, and S. Napel, "Perspective volume rendering of CT
[5] S. Geman and D. Geman, "Stochastic relaxiation, Gibbs distributions,
and MR images: Applications for endoscopic imaging," Radiology, vol.
and the Bayesian restoration of images," IEEE Trans. Pattern Anal. Ma-
199, pp. 321–330, 1996.
chine Intell., vol. 6, pp. 721–741, 1984.
[15] B. Simons, A. Morrison, R. Lev, and W. Verhoek-Oftendahl, "Relation-
[6] A. Hara, C. Johnson, J. Reed, D. Ahlquist, H. Nelson, R. Ehman, C. Mc-
ship of polyps to cancer of the large intestine," J. National Cancer Inst.,
Collough, and D. Ilstrup, "Detection of Colorectal Polyps by CT Colog-
vol. 84, pp. 962–966, 1992.
raphy: Feasibility of a novel technique," Gastroenterology, vol. 110, pp.
[16] N. V. Smirnov, "On the estimation of discrepancy between empirical
284–290, 1996.
curves of distribution for two independent samples" (in Russian), Bull.
[7] L. Hong, A. Kaufman, Y.-C. Wei, A. Viswambharan, M. Wax, and Z.
Moscow Univ., vol. 2, pp. 3–16, 1939.
Liang, "3-D virtual colonoscopy," in Proc. Biomedical Visualization, M.
Loew and N. Gershon, Eds., Atlanta, GA, 1995, pp. 26–33.
London, U.K.: Chapman & Hall, 1993.
[8] L. Hong, Z. Liang, A. Viswambharan, A. Kaufman, and M. Wax, "Re-
[18] D. J. Vining, D. Gelfand, R. Bechtold, E. Scharling, E. F. Grishaw, and
construction and visualization of 3-D models of colonic surface," IEEE
R. Shifirin, "Technical feasibility of colon imaging with helical CT and
Trans. Nucl. Sci., vol. 44, pp. 1297–1302, 1997.
virtual reality," in 1994 Ann. Meeting Amer. Roentgen Ray. Soc., New
[9] Z. Liang, F. Yang, M. Wax, J. Li, J. You, A. Kaufman, L. Hong, H. Li, and
Orleans, p. 104.
A. Viswambharan, "Inclusion of a priori information in segmentation of
[19] J. Zhang, J. W. Modestino, and D. A. Langan, "Maximum-likelihood pa-
colon lumen for 3-D virtual colonoscapy," in Conf. Rec. IEEE NSS-MIC,
rameter estimation for unsupervised stochastic model-based image seg-
Albuquerque, NM, Nov. 1997.
mentation," IEEE Trans. Image Processing, vol. 3, pp. 404–420, 1994.
[10] Y. Linde, A. Buzo, and R. M. Gray, "An algorithm for vector quantizer
[20] "Cancer facts and figures," Amer. Cancer Soc., Atlanta, GA, pt. 2, 1998.
designed," IEEE Trans. Commun., vol. 28, pp. 84–95, 1980.
[11] E. McFarland, J. Brink, J. Loh, G. Wang, V. Argiro, D. Balfe, J. Heiken,
and M. Vannier, "Visualization of colorectal polyps with spiral CTcolography: Evaluation of processing parameters with perspectivevolume rendering," Radiology, vol. 205, pp. 701–707, 1997.
Source: https://cvc.cs.stonybrook.edu/Publications/2000/CLMLLK00/file.pdf
I QUADERNI DEL MASTER Ex OSP2 e Farmacia di Comunità I necessari approfondimenti per dispensare consapevolmente medicinali innovativi Università degli Studi Ordine dei Farmacisti di Torino della Provincia di Torino Con il patrocinio di: FEDERFARMA I QUADERNI DEL MASTER Ex OSP2 e Farmacia di Comunità
Atherosclerosis Supplements 16 (2015) 12–16 Alterations of intestinal lipoprotein metabolism in diabetes mellitus and metabolic syndrome Dipartimento di Medicina Interna e Specialità Mediche, UOS Centro Arteriosclerosi Università di Roma La Sapienza, Rome, Italy Diabetes and metabolic syndrome are associated with abnormal postprandial lipoprotein metabolism, with a significant delay in the




